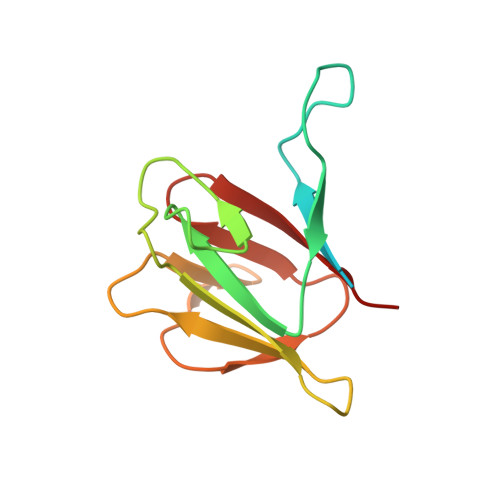
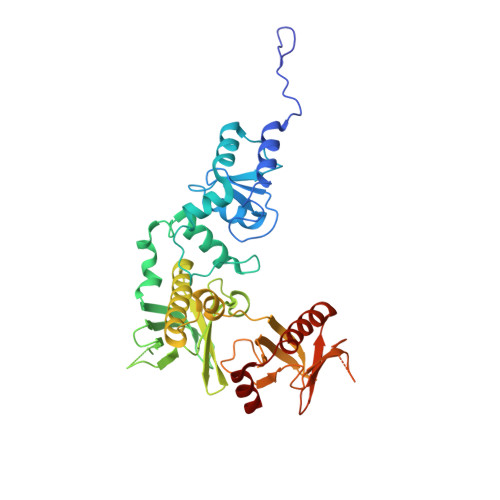
Phosphorylation-independent dual-site binding of the FHA domain of KIF13 mediates phosphoinositide transport via centaurin alpha1.
Tong, Y., Tempel, W., Wang, H., Yamada, K., Shen, L., Senisterra, G.A., MacKenzie, F., Chishti, A.H., Park, H.W.(2010) Proc Natl Acad Sci U S A 107: 20346-20351
- PubMed: 21057110
- DOI: https://doi.org/10.1073/pnas.1009008107
- Primary Citation of Related Structures:
3FEH, 3FM8, 3LJU - PubMed Abstract:
Phosphatidylinositol 3,4,5-triphosphate (PIP3) plays a key role in neuronal polarization and axon formation. PIP3-containing vesicles are transported to axon tips by the kinesin KIF13B via an adaptor protein, centaurin α1 (CENTA1). KIF13B interacts with CENTA1 through its forkhead-associated (FHA) domain. We solved the crystal structures of CENTA1 in ligand-free, KIF13B-FHA domain-bound, and PIP3 head group (IP4)-bound conformations, and the CENTA1/KIF13B-FHA/IP4 ternary complex. The first pleckstrin homology (PH) domain of CENTA1 specifically binds to PIP3, while the second binds to both PIP3 and phosphatidylinositol 3,4-biphosphate (PI(3,4)P(2)). The FHA domain of KIF13B interacts with the PH1 domain of one CENTA1 molecule and the ArfGAP domain of a second CENTA1 molecule in a threonine phosphorylation-independent fashion. We propose that full-length KIF13B and CENTA1 form heterotetramers that can bind four phosphoinositide molecules in the vesicle and transport it along the microtubule.
- Structural Genomics Consortium, University of Toronto, Toronto, ON M5G 1L7, Canada.
Organizational Affiliation: