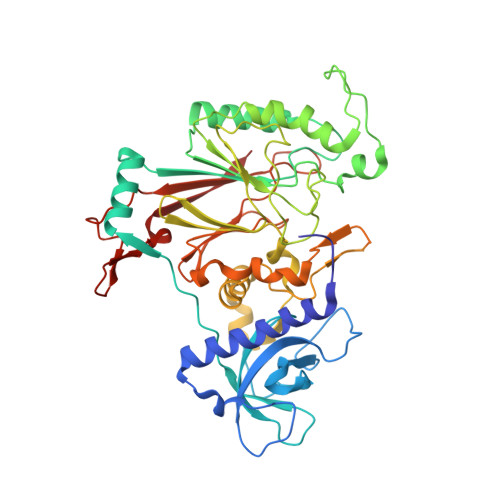
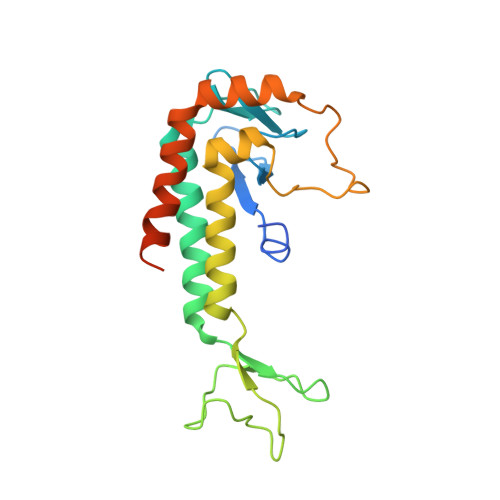
3D architecture of DNA Pol alpha reveals the functional core of multi-subunit replicative polymerases
Klinge, S., Nunez-Ramirez, R., Llorca, O., Pellegrini, L.(2009) EMBO J 28: 1978-1987
- PubMed: 19494830
- DOI: https://doi.org/10.1038/emboj.2009.150
- Primary Citation of Related Structures:
3FLO - PubMed Abstract:
Eukaryotic DNA replication requires the coordinated activity of the multi-subunit DNA polymerases: Pol alpha, Pol delta and Pol epsilon. The conserved catalytic and regulatory B subunits associate in a constitutive heterodimer that represents the functional core of all three replicative polymerases. Here, we combine X-ray crystallography and electron microscopy (EM) to describe subunit interaction and 3D architecture of heterodimeric yeast Pol alpha. The crystal structure of the C-terminal domain (CTD) of the catalytic subunit bound to the B subunit illustrates a conserved mechanism of accessory factor recruitment by replicative polymerases. The EM reconstructions of Pol alpha reveal a bilobal shape with separate catalytic and regulatory modules. Docking of the B-CTD complex in the EM reconstruction shows that the B subunit is tethered to the polymerase domain through a structured but flexible linker. Our combined findings provide a structural template for the common functional architecture of the three major replicative DNA polymerases.
- Department of Biochemistry, University of Cambridge, Cambridge, UK.
Organizational Affiliation: