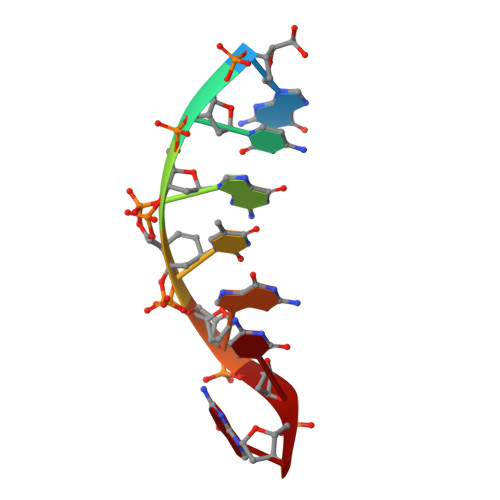
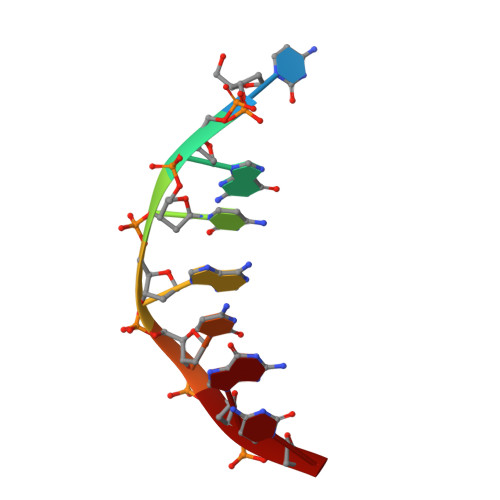
Direct observation of two cyclohexenyl (CeNA) ring conformations in duplex DNA.
Robeyns, K., Herdewijn, P., Van Meervelt, L.(2010) Artif DNA PNA XNA 1: 2-8
- PubMed: 21687521
- DOI: https://doi.org/10.4161/adna.1.1.10952
- Primary Citation of Related Structures:
3FL6 - PubMed Abstract:
Cyclohexene Nucleic Acids (CeNA), in which the 2'-deoxyribofuranose ring of the DNA building blocks is substituted by a cyclohexenyl ring, were designed as potential mimics of natural nucleic acids for antisense and, later, for siRNA applications. CeNA units, in contrast to HNA (hexitol nucleic acid) building blocks, show more flexibility at the level of the C2'-C3' bond due to the possibility of the cyclohexenyl moiety to adopt different conformations. In order to analyze the influence of CeNA residues onto the helix conformation and hydration of natural nucleic acid structures and to verify the cyclohexenyl ring conformation, a cyclohexenyl-thymine building block was incorporated into the non-self-complementary sequence d(GCG(xT)GCG)/d(CGCACGC) with (xT) a cyclohexene residue. The crystal structure of this sequence has been determined to a resolution of 1.17 Å and contains two duplexes in the asymmetric unit. The global helices belong to the B-type family and the conformations of the cyclohexenyl rings in both duplexes are different. The cyclohexene ring adopts as well the (2)H(3)-conformation (similar to C2'-endo) as the (3)H(2)-conformation (similar to C3'-endo). The crystal packing is stabilized by cobalt hexamine residues and triplet formation.
- Department of Chemistry, Biomolecular Architecture and BioMacS; Katholieke Universiteit Leuven; Leuven, Belgium.
Organizational Affiliation: