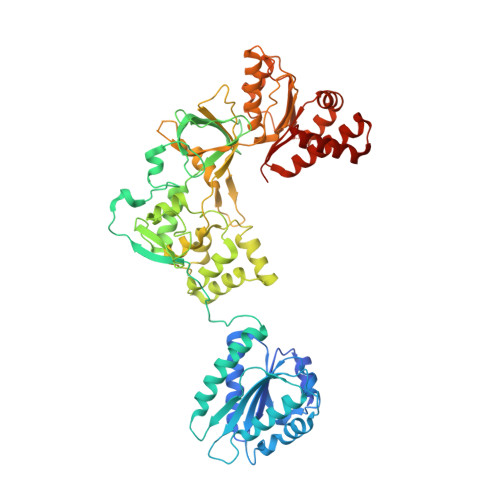
Structure of the open conformation of a functional chimeric NADPH cytochrome P450 reductase
Aigrain, L., Pompon, D., Morera, S., Truan, G.(2009) EMBO Rep 10: 742-747
- PubMed: 19483672
- DOI: https://doi.org/10.1038/embor.2009.82
- Primary Citation of Related Structures:
3FJO - PubMed Abstract:
Two catalytic domains, bearing FMN and FAD cofactors, joined by a connecting domain, compose the core of the NADPH cytochrome P450 reductase (CPR). The FMN domain of CPR mediates electron shuttling from the FAD domain to cytochromes P450. Together, both enzymes form the main mixed-function oxidase system that participates in the metabolism of endo- and xenobiotic compounds in mammals. Available CPR structures show a closed conformation, with the two cofactors in tight proximity, which is consistent with FAD-to-FMN, but not FMN-to-P450, electron transfer. Here, we report the 2.5 A resolution crystal structure of a functionally competent yeast-human chimeric CPR in an open conformation, compatible with FMN-to-P450 electron transfer. Comparison with closed structures shows a major conformational change separating the FMN and FAD cofactors from 86 A.
- Centre de Génétique Moléculaire, FRE3144, Gif-sur-Yvette, France.
Organizational Affiliation: