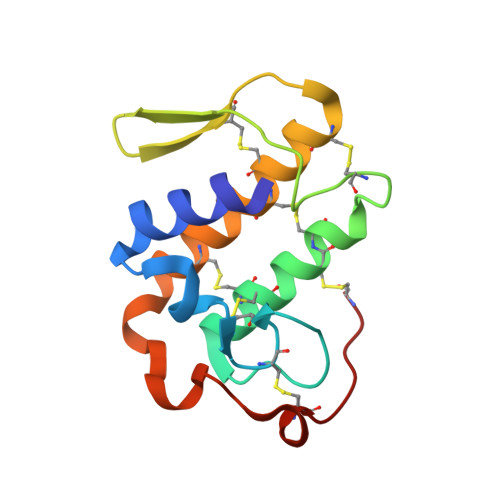
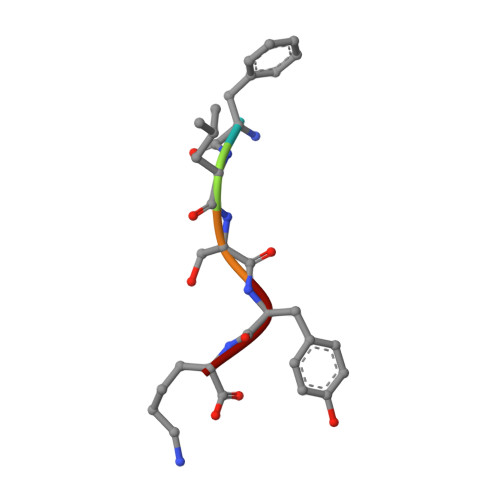
Crystal structure determination of a ternary complex of phospholipase A2 with a pentapeptide FLSYK and Ajmaline at 2.5 A resolution
Kumar, M., Kumar, S., Vikram, G., Singh, N., Sinha, M., Bhushan, A., Kaur, P., Srinivasan, A., Sharma, S., Singh, T.P.To be published.