The shaping of T cell receptor recognition by self-tolerance.
Gras, S., Burrows, S.R., Kjer-Nielsen, L., Clements, C.S., Liu, Y.C., Sullivan, L.C., Bell, M.J., Brooks, A.G., Purcell, A.W., McCluskey, J., Rossjohn, J.(2009) Immunity 30: 193-203
- PubMed: 19167249
- DOI: https://doi.org/10.1016/j.immuni.2008.11.011
- Primary Citation of Related Structures:
3FFC - PubMed Abstract:
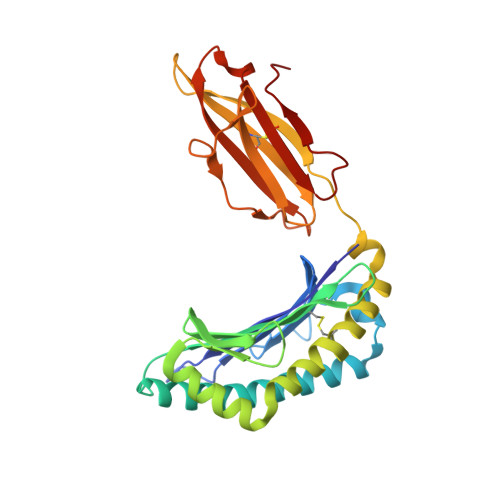
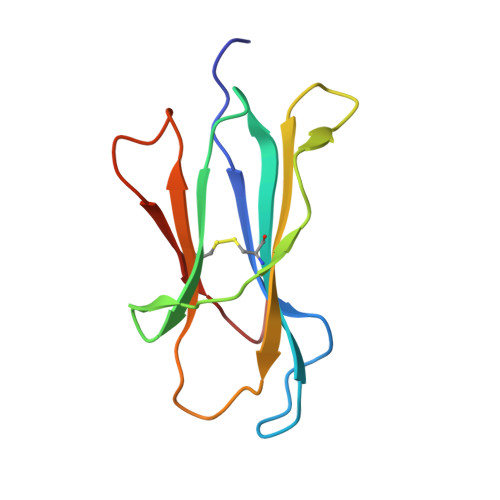
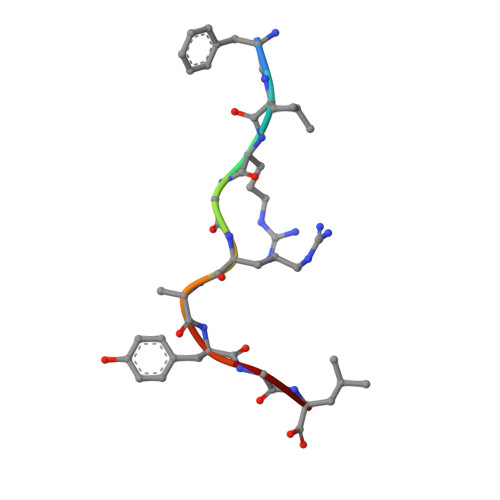
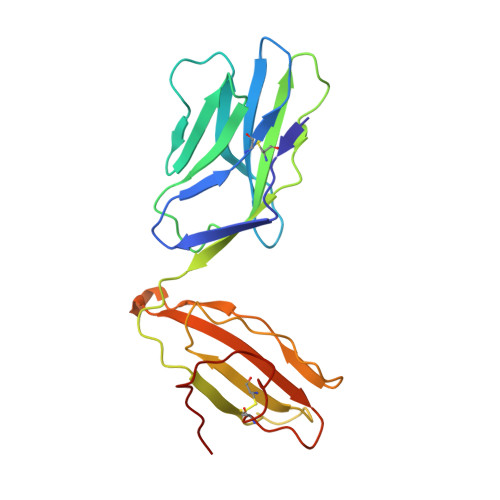
During selection of the T cell repertoire, the immune system navigates the subtle distinction between self-restriction and self-tolerance, yet how this is achieved is unclear. Here we describe how self-tolerance toward a trans-HLA (human leukocyte antigen) allotype shapes T cell receptor (TCR) recognition of an Epstein-Barr virus (EBV) determinant (FLRGRAYGL). The recognition of HLA-B8-FLRGRAYGL by two archetypal TCRs was compared. One was a publicly selected TCR, LC13, that is alloreactive with HLA-B44; the other, CF34, lacks HLA-B44 reactivity because it arises when HLA-B44 is coinherited in trans with HLA-B8. Whereas the alloreactive LC13 TCR docked at the C terminus of HLA-B8-FLRGRAYGL, the CF34 TCR docked at the N terminus of HLA-B8-FLRGRAYGL, which coincided with a polymorphic region between HLA-B8 and HLA-B44. The markedly contrasting footprints of the LC13 and CF34 TCRs provided a portrait of how self-tolerance shapes the specificity of TCRs selected into the immune repertoire.
- Department of Biochemistry and Molecular Biology, The Protein Crystallography Unit, Monash University, Clayton, Victoria, Australia.
Organizational Affiliation: