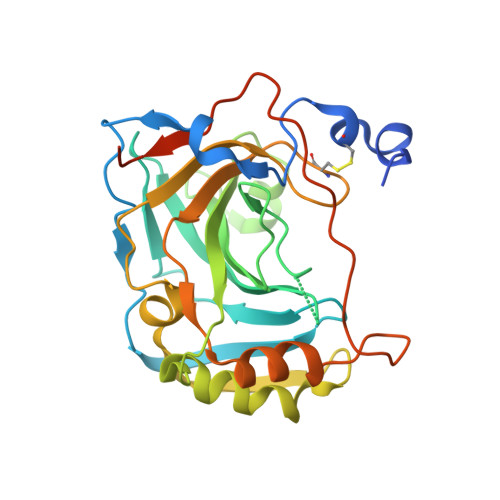
Crystal structure of the secretory isozyme of mammalian carbonic anhydrases CA VI: implications for biological assembly and inhibitor development
Pilka, E.S., Kochan, G., Oppermann, U., Yue, W.W.(2012) Biochem Biophys Res Commun 419: 485-489
- PubMed: 22366092
- DOI: https://doi.org/10.1016/j.bbrc.2012.02.038
- Primary Citation of Related Structures:
3FE4 - PubMed Abstract:
Zn(2+)-dependent carbonic anhydrases (CA) catalyse the reversible hydration of carbon dioxide to bicarbonate and participate in diverse physiological processes, hence having manifold therapeutic potentials. Among the 15 human CAs with wide-ranging sub-cellular localisation and kinetic properties, CA VI is the only secretory isoform. The 1.9Å crystal structure of the human CA VI catalytic domain reveals a prototypical mammalian CA fold, and a novel dimeric arrangement as compared to previously-reported CA structures. The active site cavity contains a cluster of non-conserved residues that may be involved in ligand binding and have significant implications for developing the next-generation of isoform-specific inhibitors.
- Structural Genomics Consortium, Oxford, UK.
Organizational Affiliation: