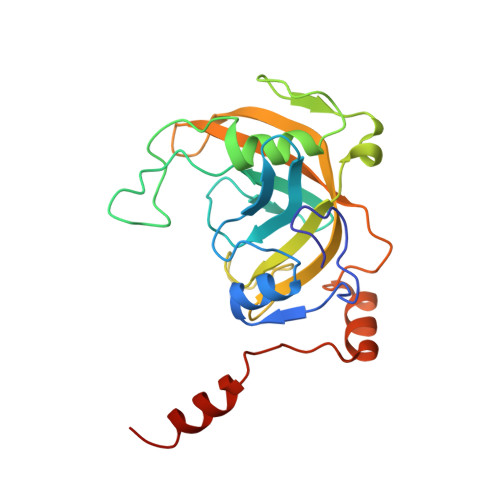
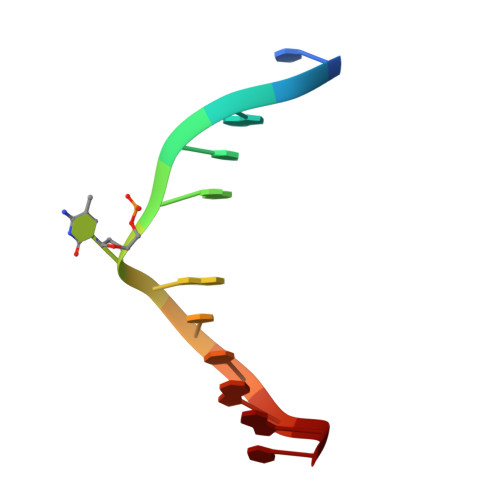
UHRF1, a modular multi-domain protein, regulates replication-coupled crosstalk between DNA methylation and histone modifications.
Hashimoto, H., Horton, J.R., Zhang, X., Cheng, X.(2009) Epigenetics 4: 8-14
- PubMed: 19077538
- DOI: https://doi.org/10.4161/epi.4.1.7370
- Primary Citation of Related Structures:
3F8I, 3F8J, 3FDE - PubMed Abstract:
Cytosine methylation in DNA is a major epigenetic signal, and plays a central role in propagating chromatin status during cell division. However the mechanistic links between DNA methylation and histone methylation are poorly understood. A multi-domain protein UHRF1 (ubiquitin-like, containing PHD and RING finger domains 1) is required for DNA CpG maintenance methylation at replication forks, and mouse UHRF1-null cells show enhanced susceptibility to DNA replication arrest and DNA damaging agents. Recent data demonstrated that the SET and RING associated (SRA) domain of UHRF1 binds hemimethylated CpG and flips 5-methylcytosine out of the DNA helix, whereas its tandom tudor domain and PHD domain bind the tail of histone H3 in a highly methylation sensitive manner. We hypothesize that UHRF1 brings the two components (histones and DNA) carrying appropriate markers (on the tails of H3 and hemimethylated CpG sites) ready to be assembled into a nucleosome after replication.
- Department of Biochemistry, Emory University School of Medicine, Atlanta, Georgia 30322, USA.
Organizational Affiliation: