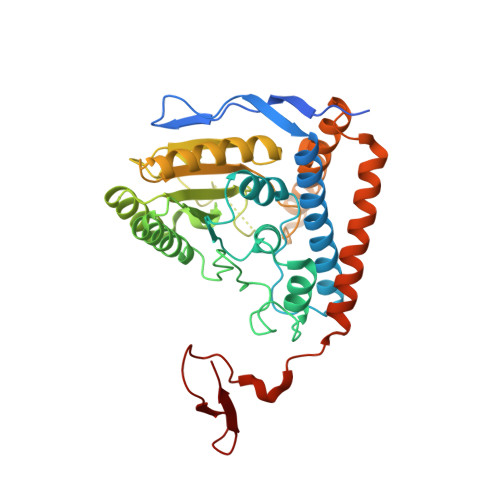
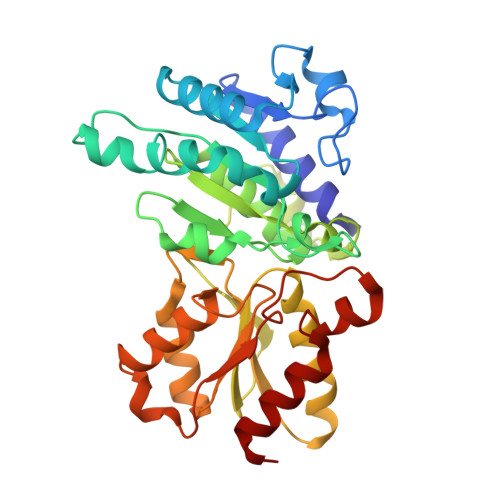
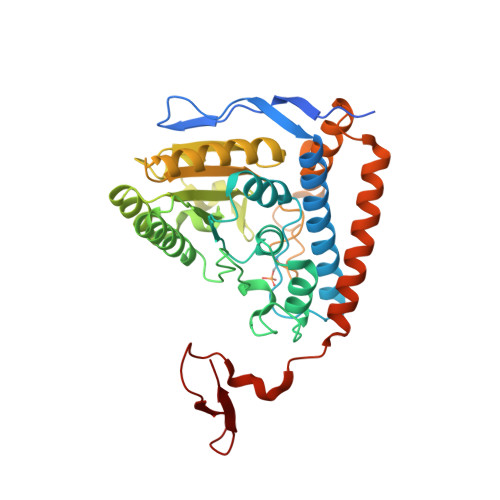
Structural basis for inactivation of the human pyruvate dehydrogenase complex by phosphorylation: role of disordered phosphorylation loops.
Kato, M., Wynn, R.M., Chuang, J.L., Tso, S.C., Machius, M., Li, J., Chuang, D.T.(2008) Structure 16: 1849-1859
- PubMed: 19081061
- DOI: https://doi.org/10.1016/j.str.2008.10.010
- Primary Citation of Related Structures:
3EXE, 3EXF, 3EXG, 3EXH, 3EXI - PubMed Abstract:
We report the crystal structures of the phosporylated pyruvate dehydrogenase (E1p) component of the human pyruvate dehydrogenase complex (PDC). The complete phosphorylation at Ser264-alpha (site 1) of a variant E1p protein was achieved using robust pyruvate dehydrogenase kinase 4 free of the PDC core. We show that unlike its unmodified counterpart, the presence of a phosphoryl group at Ser264-alpha prevents the cofactor thiamine diphosphate-induced ordering of the two loops carrying the three phosphorylation sites. The disordering of these phosphorylation loops is caused by a previously unrecognized steric clash between the phosphoryl group at site 1 and a nearby Ser266-alpha, which nullifies a hydrogen-bonding network essential for maintaining the loop conformations. The disordered phosphorylation loops impede the binding of lipoyl domains of the PDC core to E1p, negating the reductive acetylation step. This results in the disruption of the substrate channeling in the PDC, leading to the inactivation of this catalytic machine.
- Department of Internal Medicine, University of Texas Southwestern Medical Center, Dallas, TX 75390-9038, USA.
Organizational Affiliation: