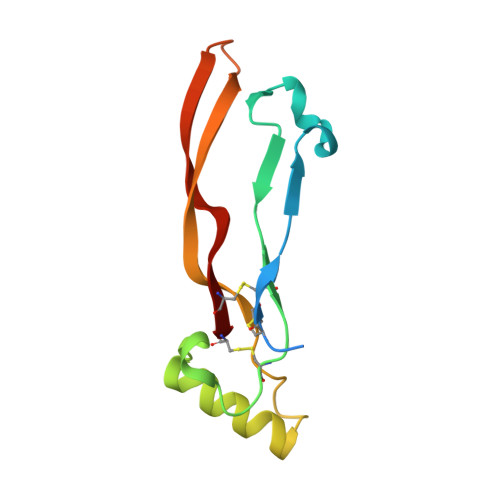
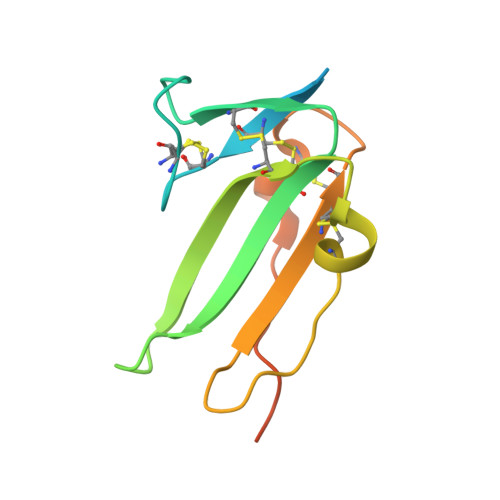
Crystal structure analysis reveals a spring-loaded latch as molecular mechanism for GDF-5-type I receptor specificity.
Kotzsch, A., Nickel, J., Seher, A., Sebald, W., Muller, T.D.(2009) EMBO J 28: 937-947
- PubMed: 19229295
- DOI: https://doi.org/10.1038/emboj.2009.37
- Primary Citation of Related Structures:
3EVS - PubMed Abstract:
Dysregulation of growth and differentiation factor 5 (GDF-5) signalling, a member of the TGF-beta superfamily, is strongly linked to skeletal malformation. GDF-5-mediated signal transduction involves both BMP type I receptors, BMPR-IA and BMPR-IB. However, mutations in either GDF-5 or BMPR-IB lead to similar phenotypes, indicating that in chondrogenesis GDF-5 signalling seems to be exclusively mediated through BMPR-IB. Here, we present structural insights into the GDF-5:BMPR-IB complex revealing how binding specificity for BMPR-IB is generated on a molecular level. In BMPR-IB, a loop within the ligand-binding epitope functions similar to a latch allowing high-affinity binding of GDF-5. In BMPR-IA, this latch is in a closed conformation leading to steric repulsion. The new structural data now provide also a molecular basis of how phenotypically relevant missense mutations in GDF-5 might impair receptor binding and activation.
- Lehrstuhl für Botanik I-Molekulare Pflanzenphysiologie und Biophysik, Julius-von-Sachs-Institut für Biowissenschaften (Biozentrum) der Universität Würzburg, Würzburg, Germany.
Organizational Affiliation: