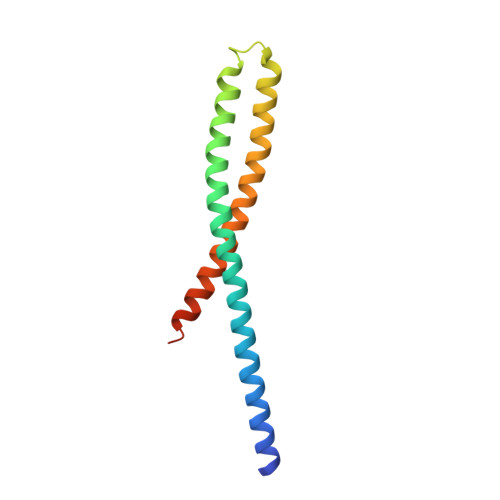Crystal Structure of FadA Adhesin from Fusobacterium nucleatum Reveals a Novel Oligomerization Motif, the Leucine Chain.
Nithianantham, S., Xu, M., Yamada, M., Ikegami, A., Shoham, M., Han, Y.W.(2009) J Biological Chem 284: 3865-3872
- PubMed: 18996848
- DOI: https://doi.org/10.1074/jbc.M805503200
- Primary Citation of Related Structures:
3ETW, 3ETX, 3ETY, 3ETZ - PubMed Abstract:
Many bacterial appendages have filamentous structures, often composed of repeating monomers assembled in a head-to-tail manner. The mechanisms of such linkages vary. We report here a novel protein oligomerization motif identified in the FadA adhesin from the Gram-negative bacterium Fusobacterium nucleatum. The 2.0 angstroms crystal structure of the secreted form of FadA (mFadA) reveals two antiparallel alpha-helices connected by an intervening 8-residue hairpin loop. Leucine-leucine contacts play a prominent dual intra- and intermolecular role in the structure and function of FadA. First, they comprise the main association between the two helical arms of the monomer; second, they mediate the head-to-tail association of monomers to form the elongated polymers. This leucine-mediated filamentous assembly of FadA molecules constitutes a novel structural motif termed the "leucine chain." The essential role of these residues in FadA is corroborated by mutagenesis of selected leucine residues, which leads to the abrogation of oligomerization, filament formation, and binding to host cells.
- Department of Biochemistry, Case Western Reserve University, Cleveland, Ohio 44106, USA.
Organizational Affiliation: