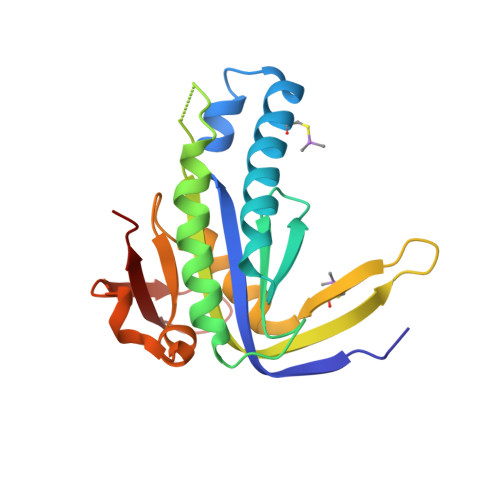
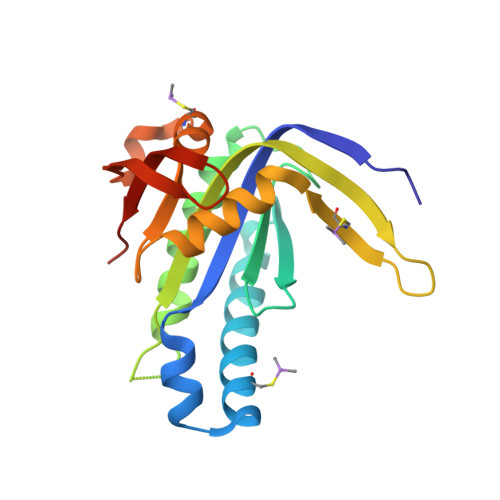
The crystal structure of the catalytic domain of a eukaryotic guanylate cyclase.
Winger, J.A., Derbyshire, E.R., Lamers, M.H., Marletta, M.A., Kuriyan, J.(2008) BMC Struct Biol 8: 42-42
- PubMed: 18842118
- DOI: https://doi.org/10.1186/1472-6807-8-42
- Primary Citation of Related Structures:
3ET6 - PubMed Abstract:
Soluble guanylate cyclases generate cyclic GMP when bound to nitric oxide, thereby linking nitric oxide levels to the control of processes such as vascular homeostasis and neurotransmission. The guanylate cyclase catalytic module, for which no structure has been determined at present, is a class III nucleotide cyclase domain that is also found in mammalian membrane-bound guanylate and adenylate cyclases. We have determined the crystal structure of the catalytic domain of a soluble guanylate cyclase from the green algae Chlamydomonas reinhardtii at 2.55 A resolution, and show that it is a dimeric molecule. Comparison of the structure of the guanylate cyclase domain with the known structures of adenylate cyclases confirms the close similarity in architecture between these two enzymes, as expected from their sequence similarity. The comparison also suggests that the crystallized guanylate cyclase is in an inactive conformation, and the structure provides indications as to how activation might occur. We demonstrate that the two active sites in the dimer exhibit positive cooperativity, with a Hill coefficient of approximately 1.5. Positive cooperativity has also been observed in the homodimeric mammalian membrane-bound guanylate cyclases. The structure described here provides a reliable model for functional analysis of mammalian guanylate cyclases, which are closely related in sequence.
- Department of Molecular and Cell Biology, University of California, Berkeley, CA, USA. wingerj@berkeley.edu
Organizational Affiliation: