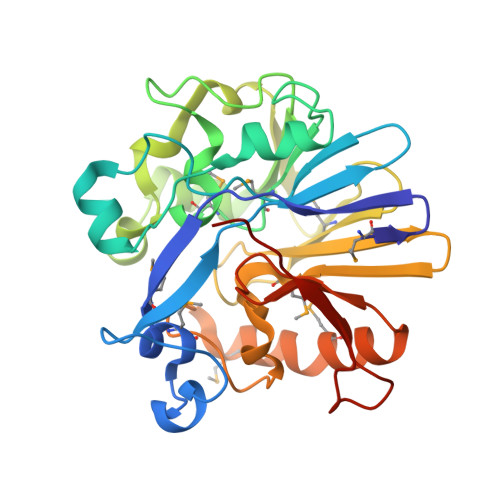
Crystal structure of a probable metal-dependent hydrolase from Staphylococcus aureus. Northeast Structural Genomics target ZR314
Seetharaman, J., Chen, Y., Wang, H., Janjua, H., Foote, E.L., Xiao, R., Nair, R., Everett, J.K., Acton, T.B., Rost, B., Montelione, G.T., Tong, L., Hunt, J.F.To be published.