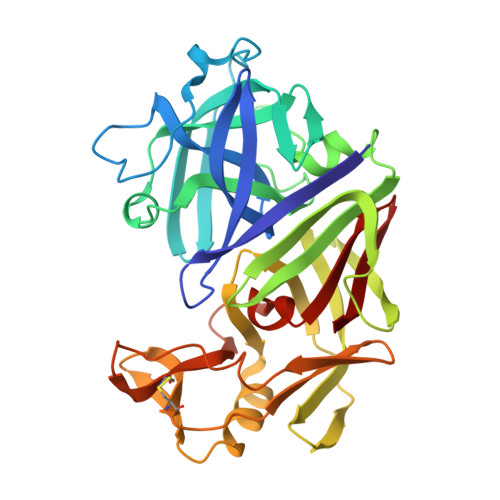
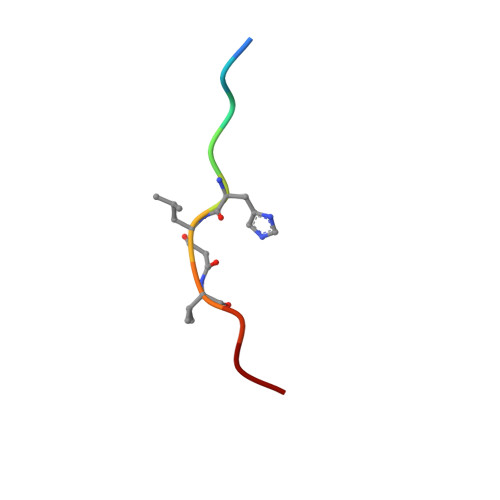
X-ray-crystallographic studies of complexes of pepstatin A and a statine-containing human renin inhibitor with endothiapepsin.
Bailey, D., Cooper, J.B., Veerapandian, B., Blundell, T.L., Atrash, B., Jones, D.M., Szelke, M.(1993) Biochem J 289 ( Pt 2): 363-371
- PubMed: 8424781
- DOI: https://doi.org/10.1042/bj2890363
- Primary Citation of Related Structures:
3ER5 - PubMed Abstract:
H-189, a synthetic human renin inhibitor, and pepstatin A, a naturally occurring inhibitor of aspartic proteinases, have been co-crystallized with the fungal aspartic proteinase endothiapepsin (EC 3.4.23.6). H-189 [Pro-His-Pro-Phe-His-Sta-(statyl)-Val-Ile-His-Lys] is an analogue of human angiotensinogen. Pepstatin A [Iva(isovaleryl)-Val-Val-Sta-Ala-Sta] is a blocked pentapeptide which inhibits many aspartic proteinases. The structures of the complexes have been determined by X-ray diffraction and refined to crystallographic R-factors of 0.15 and 0.16 at resolutions of 0.18 nm (1.8 A) and 0.2 nm (2.0 A) respectively. H-189 is in an extended conformation, in which the statine residue is a dipeptide analogue of P1 and P'1 as indicated by the conformation and network of contacts and hydrogen bonds. Pepstatin A has an extended conformation to the P'2 alanine residue, but the leucyl side chain of the terminal statine residue binds back into the S'1 subsite, and an inverse gamma-turn occurs between P'1 and P'3. The hydroxy moiety of the statine at P1 in both complexes displaces the solvent molecule that hydrogen-bonds with the catalytic aspartate residues (32 and 215) in the native enzyme. Solvent molecules originally present in the native structure at the active site are displaced on inhibitor binding (12 when pepstatin A binds; 16 when H-189 binds).
- Department of Crystallography, Birkbeck College, University of London, U.K.
Organizational Affiliation: