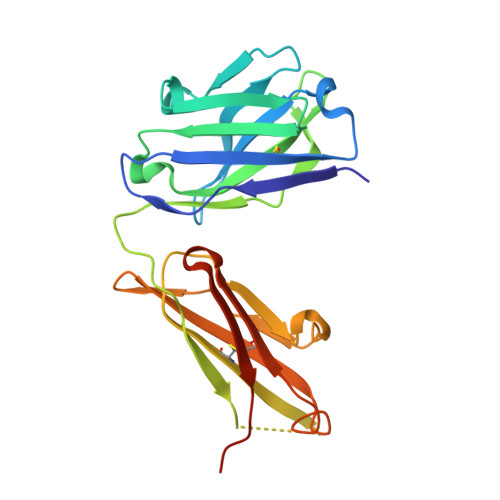
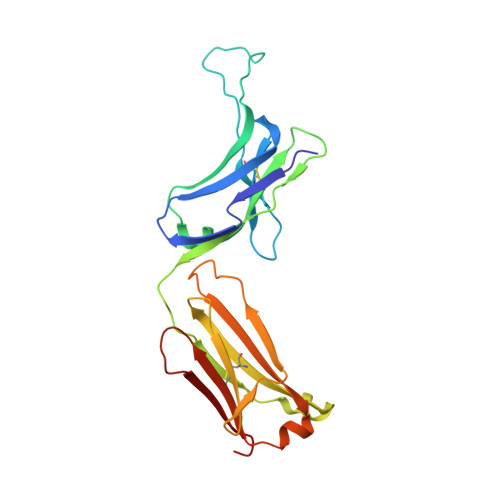
An antibody loop replacement design feasibility study and a loop-swapped dimer structure.
Clark, L.A., Boriack-Sjodin, P.A., Day, E., Eldredge, J., Fitch, C., Jarpe, M., Miller, S., Li, Y., Simon, K., van Vlijmen, H.W.(2009) Protein Eng Des Sel 22: 93-101
- PubMed: 19074157
- DOI: https://doi.org/10.1093/protein/gzn072
- Primary Citation of Related Structures:
3EOT - PubMed Abstract:
A design approach was taken to investigate the feasibility of replacing single complementarity determining region (CDR) antibody loops. This approach may complement simpler mutation-based strategies for rational antibody design by expanding conformation space. Enormous crystal structure diversity is available, making CDR loops logical targets for structure-based design. A detailed analysis for the L1 loop shows that each loop length takes a distinct conformation, thereby allowing control on a length scale beyond that accessible to simple mutations. The L1 loop in the anti-VLA1 antibody was replaced with the L2 loop residues longer in an attempt to add an additional hydrogen bond and fill space on the antibody-antigen interface. The designs expressed well, but failed to improve affinity. In an effort to learn more, one design was crystallized and data were collected at 1.9 A resolution. The designed L1 loop takes the qualitatively desired conformation; confirming that loop replacement by design is feasible. The crystal structure also shows that the outermost loop (residues Leu51-Ser68) is domain swapped with another monomer. Tryptophan fluorescence measurements were used to monitor unfolding as a function of temperature and indicate that the loop involved in domain swapping does not unfold below 60 degrees C. The domain-swapping is not directly responsible for the affinity loss, but is likely a side-effect of the structural instability which may contribute to affinity loss. A second round of design was successful in eliminating the dimerization through mutation of a residue (Leu51Ser) at the joint of the domain-swapped loop.
- Biogen Idec Inc., Cambridge, MA 02142, USA. louie@alumni.northwestern.edu
Organizational Affiliation: