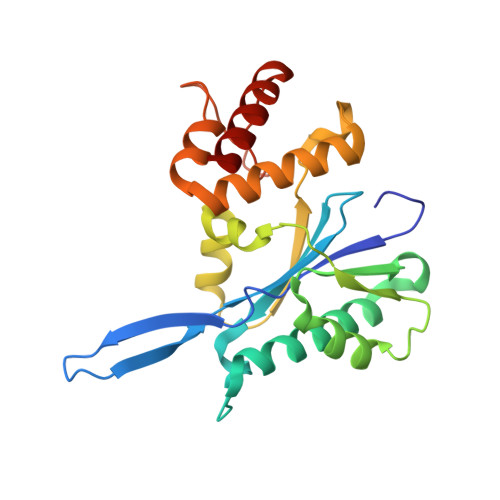
Structural elucidation of a PRP8 core domain from the heart of the spliceosome.
Ritchie, D.B., Schellenberg, M.J., Gesner, E.M., Raithatha, S.A., Stuart, D.T., Macmillan, A.M.(2008) Nat Struct Mol Biol 15: 1199-1205
- PubMed: 18836455
- DOI: https://doi.org/10.1038/nsmb.1505
- Primary Citation of Related Structures:
3ENB - PubMed Abstract:
The spliceosome is a complex ribonucleoprotein (RNP) particle containing five RNAs and more than 100 associated proteins. One of these proteins, PRP8, has been shown to interact directly with the splice sites and branch region of precursor-mRNAs (pre-mRNAs) and spliceosomal RNAs associated with catalysis of the two steps of splicing. The 1.85-A X-ray structure of the core of PRP8 domain IV, implicated in key spliceosomal interactions, reveals a bipartite structure that includes the presence of an RNase H fold linked to a five-helix assembly. Analysis of mutant yeast alleles and cross-linking results in the context of this structure, coupled with RNA binding studies, suggests that domain IV forms a surface that interacts directly with the RNA structures at the catalytic core of the spliceosome.
- Department of Biochemistry, 474 Medical Sciences Building, University of Alberta, Edmonton, Alberta T6G2H7, Canada.
Organizational Affiliation: