Optimization of in vivo activity of a bifunctional homing endonuclease and maturase reverses evolutionary degradation.
Takeuchi, R., Certo, M., Caprara, M.G., Scharenberg, A.M., Stoddard, B.L.(2009) Nucleic Acids Res 37: 877-890
- PubMed: 19103658
- DOI: https://doi.org/10.1093/nar/gkn1007
- Primary Citation of Related Structures:
3EH8 - PubMed Abstract:
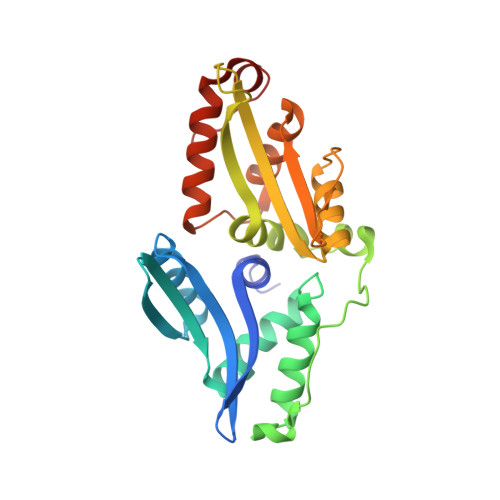
The LAGLIDADG homing endonuclease (LHE) I-AniI has adopted an extremely efficient secondary RNA splicing activity that is beneficial to its host, balanced against inefficient DNA cleavage. A selection experiment identified point mutations in the enzyme that act synergistically to improve endonuclease activity. The amino-acid substitutions increase target affinity, alter the thermal cleavage profile and significantly increase targeted recombination in transfected cells. The RNA splicing activity is not affected by these mutations. The improvement in DNA cleavage activity is largely focused on one of the enzyme's two active sites, corresponding to a rearrangement of a lysine residue hypothesized to act as a general base. Most of the constructs isolated in the screen contain one or more mutations that revert an amino-acid identity to a residue found in one or more close homologues of I-AniI. This implies that mutations that have previously reduced the endonuclease activity of I-AniI are identified and reversed, sometimes in combination with additional 'artificial' mutations, to optimize its in vivo activity.
- Division of Basic Sciences, Fred Hutchinson Cancer Research Center, Seattle, WA 98109, USA.
Organizational Affiliation: