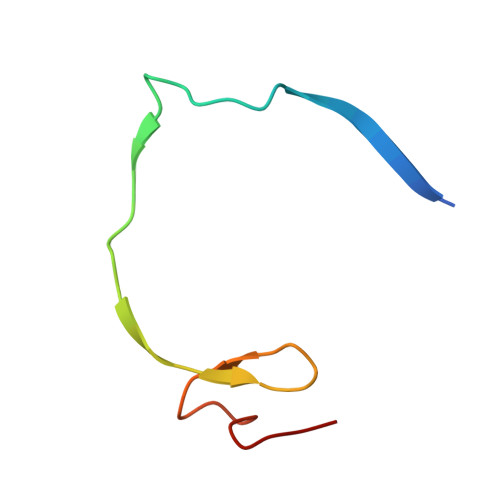
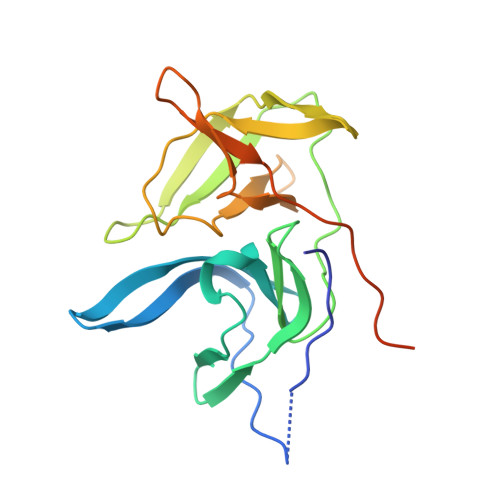
Structure of West Nile virus NS3 protease: ligand stabilization of the catalytic conformation
Robin, G., Chappell, K., Stoermer, M.J., Hu, S.-H., Young, P.R., Fairlie, D.P., Martin, J.L.(2009) J Mol Biology 385: 1568-1577
- PubMed: 19059417
- DOI: https://doi.org/10.1016/j.jmb.2008.11.026
- Primary Citation of Related Structures:
3E90 - PubMed Abstract:
Over the last decade, West Nile virus has spread rapidly via mosquito transmission from infected migratory birds to humans. One potential therapeutic approach to treating infection is to inhibit the virally encoded serine protease that is essential for viral replication. Here we report the crystal structure of the viral NS3 protease tethered to its essential NS2B cofactor and bound to a potent substrate-based tripeptide inhibitor, 2-naphthoyl-Lys-Lys-Arg-H (K(i)=41 nM), capped at the N-terminus by 2-naphthoyl and capped at the C-terminus by aldehyde. An important and unexpected feature of this structure is the presence of two conformations of the catalytic histidine suggesting a role for ligand stabilization of the catalytically competent His conformation. Analysis of other West Nile virus NS3 protease structures and related serine proteases supports this hypothesis, suggesting that the common catalytic mechanism involves an induced-fit mechanism.
- Institute for Molecular Bioscience, The University of Queensland, Brisbane, Queensland, Australia.
Organizational Affiliation: