Different vaccine vectors delivering the same antigen elicit CD8+ T cell responses with distinct clonotype and epitope specificity
Honda, M., Wang, R., Kong, W.P., Kanekiyo, M., Akahata, W., Xu, L., Matsuo, K., Natarajan, K., Robinson, H., Asher, T.E., Price, D.A., Douek, D.C., Margulies, D.H., Nabel, G.J.(2009) J Immunol 183: 2425-2434
- PubMed: 19620307
- DOI: https://doi.org/10.4049/jimmunol.0900581
- Primary Citation of Related Structures:
3E6F, 3E6H - PubMed Abstract:
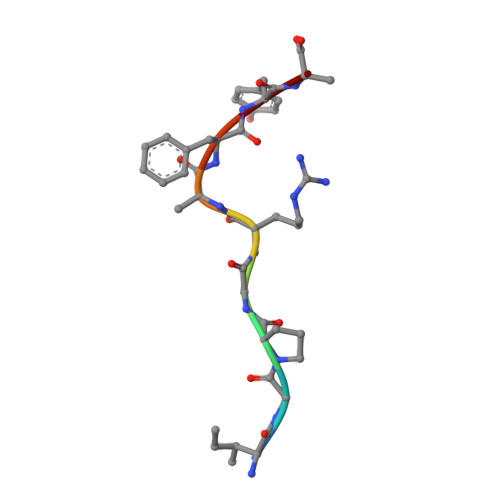
Prime-boost immunization with gene-based vectors has been developed to generate more effective vaccines for AIDS, malaria, and tuberculosis. Although these vectors elicit potent T cell responses, the mechanisms by which they stimulate immunity are not well understood. In this study, we show that immunization by a single gene product, HIV-1 envelope, with alternative vector combinations elicits CD8(+) cells with different fine specificities and kinetics of mobilization. Vaccine-induced CD8(+) T cells recognized overlapping third V region loop peptides. Unexpectedly, two anchor variants bound H-2D(d) better than the native sequences, and clones with distinct specificities were elicited by alternative vectors. X-ray crystallography revealed major differences in solvent exposure of MHC-bound peptide epitopes, suggesting that processed HIV-1 envelope gave rise to MHC-I/peptide conformations recognized by distinct CD8(+) T cell populations. These findings suggest that different gene-based vectors generate peptides with alternative conformations within MHC-I that elicit distinct T cell responses after vaccination.
- Vaccine Research Center, National Institute of Allergy and Infectious Diseases, National Institutes of Health, Bethesda, MD 20892, USA.
Organizational Affiliation: