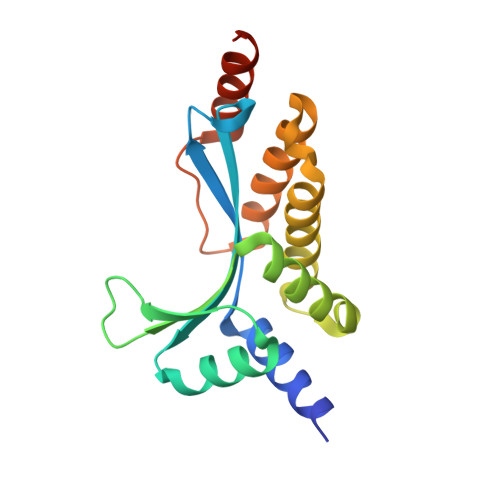
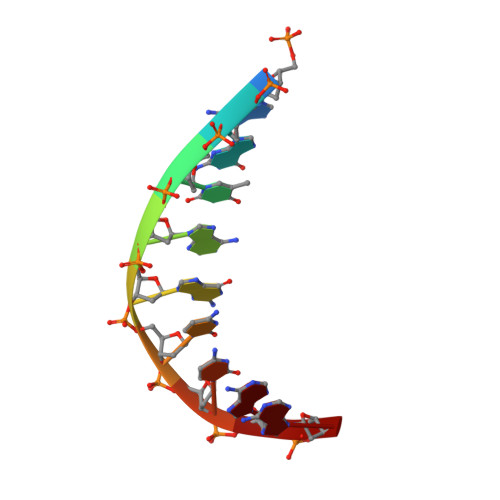
Recognition of a common rDNA target site in archaea and eukarya by analogous LAGLIDADG and His-Cys box homing endonucleases
Nomura, N., Nomura, Y., Sussman, D., Klein, D., Stoddard, B.L.(2008) Nucleic Acids Res 36: 6988-6998
- PubMed: 18984620
- DOI: https://doi.org/10.1093/nar/gkn846
- Primary Citation of Related Structures:
3E54 - PubMed Abstract:
The presence of a homing endonuclease gene (HEG) within a microbial intron or intein empowers the entire element with the ability to invade genomic targets. The persistence of a homing endonuclease lineage depends in part on conservation of its DNA target site. One such rDNA sequence has been invaded both in archaea and in eukarya, by LAGLIDADG and His-Cys box homing endonucleases, respectively. The bases encoded by this target include a universally conserved ribosomal structure, termed helix 69 (H69) in the large ribosomal subunit. This region forms the 'B2a' intersubunit bridge to the small ribosomal subunit, contacts bound tRNA in the A- and P-sites, and acts as a trigger for ribosome disassembly through its interactions with ribosome recycling factor. We have determined the DNA-bound structure and specificity profile of an archaeal LAGLIDADG homing endonuclease (I-Vdi141I) that recognizes this target site, and compared its specificity with the analogous eukaryal His-Cys box endonuclease I-PpoI. These homodimeric endonuclease scaffolds have arrived at similar specificity profiles across their common biological target and analogous solutions to the problem of accommodating conserved asymmetries within the DNA sequence, but with differences at individual base pairs that are fine-tuned to the sequence conservation of archaeal versus eukaryal ribosomes.
- Iwata Human Receptor Crystallography Project, ERATO, Japan Science and Technology Agency, Kyoto, Japan.
Organizational Affiliation: