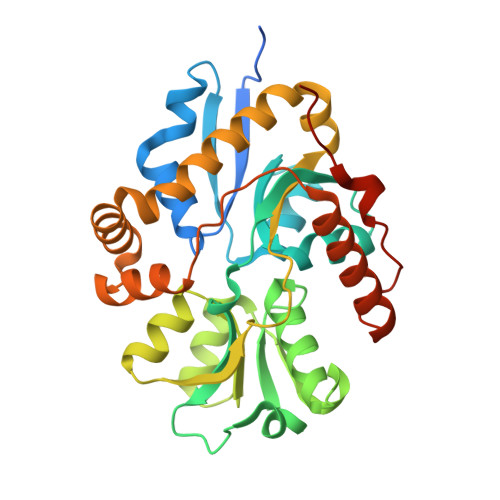
Crystal structure of the alkanesulfonate binding protein (SsuA) from the phytopathogenic bacteria Xanthomonas axonopodis pv. citri bound to HEPES
Balan, A., Araujo, F.T., Sanches, M., Chirgadze, D.Y., Blundell, T.L., Barbosa, J.A.R.G.To be published.