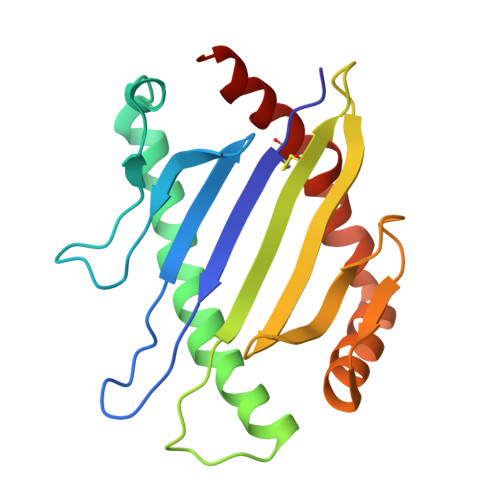
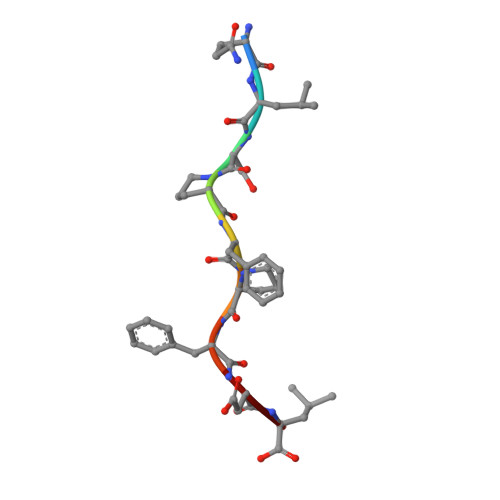
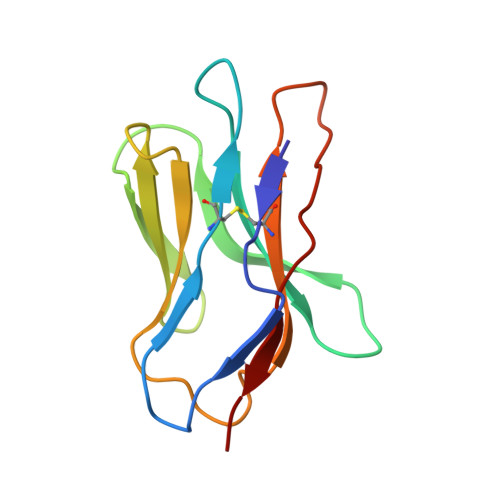
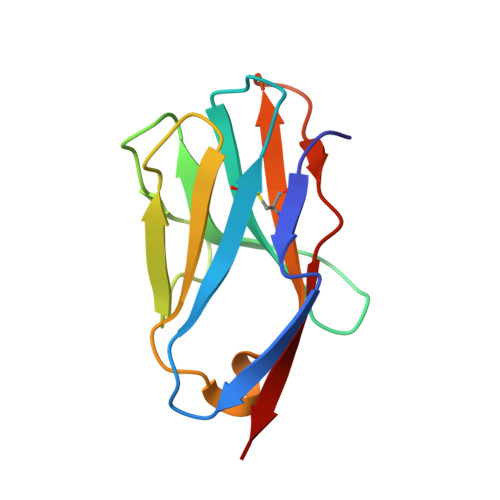
Distinct CDR3 Conformations in TCRs Determine the Level of Cross-Reactivity for Diverse Antigens, but Not the Docking Orientation
Jones, L.L., Colf, L.A., Stone, J.D., Garcia, K.C., Kranz, D.M.(2008) J Immunol 181: 6255-6264
- PubMed: 18941216
- DOI: https://doi.org/10.4049/jimmunol.181.9.6255
- Primary Citation of Related Structures:
3E2H, 3E3Q - PubMed Abstract:
T cells are known to cross-react with diverse peptide MHC Ags through their alphabeta TCR. To explore the basis of such cross-reactivity, we examined the 2C TCR that recognizes two structurally distinct ligands, SIY-K(b) and alloantigen QL9-L(d). In this study we characterized the cross-reactivity of several high-affinity 2C TCR variants that contained mutations only in the CDR3alpha loop. Two of the TCR lost their ability to cross-react with the reciprocal ligand (SIY-K(b)), whereas another TCR (m67) maintained reactivity with both ligands. Crystal structures of four of the TCRs in complex with QL9-L(d) showed that CDR1, CDR2, and CDR3beta conformations and docking orientations were remarkably similar. Although the CDR3alpha loop of TCR m67 conferred a 2000-fold higher affinity for SIY-K(b), the TCR maintained the same docking angle on QL9-L(d) as the 2C TCR. Thus, CDR3alpha dictated the affinity and level of cross-reactivity, yet it did so without affecting the conserved docking orientation.
- Department of Biochemistry, University of Illinois, Urbana, IL 61801, USA.
Organizational Affiliation: