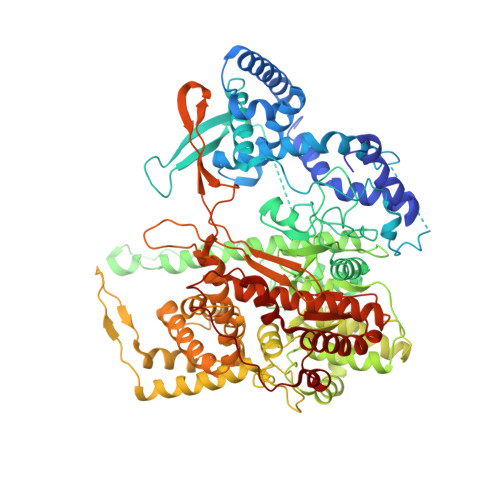
The structure of a transcribing t7 RNA polymerase in transition from initiation to elongation
Durniak, K.J., Bailey, S., Steitz, T.A.(2008) Science 322: 553-557
- PubMed: 18948533
- DOI: https://doi.org/10.1126/science.1163433
- Primary Citation of Related Structures:
3E2E, 3E3J - PubMed Abstract:
Structural studies of the T7 bacteriophage DNA-dependent RNA polymerase (T7 RNAP) have shown that the conformation of the amino-terminal domain changes substantially between the initiation and elongation phases of transcription, but how this transition is achieved remains unclear. We report crystal structures of T7 RNAP bound to promoter DNA containing either a 7- or an 8-nucleotide (nt) RNA transcript that illuminate intermediate states along the transition pathway. The amino-terminal domain comprises the C-helix subdomain and the promoter binding domain (PBD), which consists of two segments separated by subdomain H. The structures of the intermediate complex reveal that the PBD and the bound promoter rotate by approximately 45 degrees upon synthesis of an 8-nt RNA transcript. This allows the promoter contacts to be maintained while the active site is expanded to accommodate a growing heteroduplex. The C-helix subdomain moves modestly toward its elongation conformation, whereas subdomain H remains in its initiation- rather than its elongation-phase location, more than 70 angstroms away.
- Department of Molecular Biophysics and Biochemistry, Yale University, 266 Whitney Avenue, New Haven, CT 06520-8114, USA.
Organizational Affiliation: