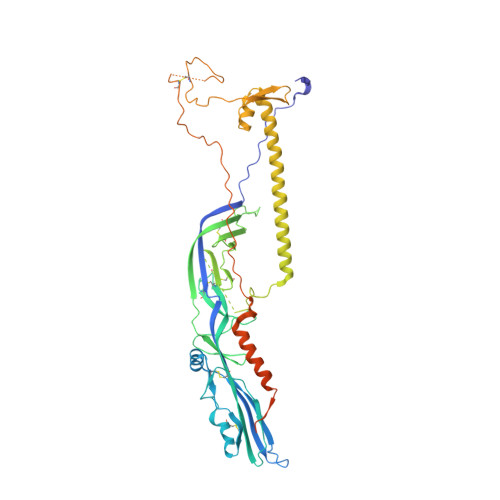
The postfusion structure of baculovirus gp64 supports a unified view of viral fusion machines.
Kadlec, J., Loureiro, S., Abrescia, N.G., Stuart, D.I., Jones, I.M.(2008) Nat Struct Mol Biol 15: 1024-1030
- PubMed: 18776902
- DOI: https://doi.org/10.1038/nsmb.1484
- Primary Citation of Related Structures:
3DUZ - PubMed Abstract:
Viral fusion proteins mediate the merger of host and viral membranes during cell entry for all enveloped viruses. Baculovirus glycoprotein gp64 (gp64) is unusual in promoting entry into both insect and mammalian cells and is distinct from established class I and class II fusion proteins. We report the crystal structure of its postfusion form, which explains a number of gp64's biological properties including its cellular promiscuity, identifies the fusion peptides and shows it to be the third representative of a new class (III) of fusion proteins with unexpected structural homology with vesicular stomatitis virus G and herpes simplex virus type 1 gB proteins. We show that domains of class III proteins have counterparts in both class I and II proteins, suggesting that all these viral fusion machines are structurally more related than previously thought.
- Division of Structural Biology, Wellcome Trust Centre for Human Genetics, University of Oxford, Roosevelt Drive, Oxford OX3 7BN, UK.
Organizational Affiliation: