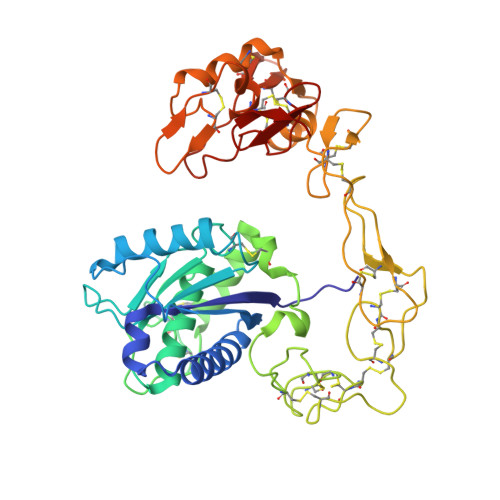
The three-dimensional structure of bothropasin, the main hemorrhagic factor from Bothrops jararaca venom: Insights for a new classification of snake venom metalloprotease subgroups.
Muniz, J.R., Ambrosio, A.L., Selistre-de-Araujo, H.S., Cominetti, M.R., Moura-da-Silva, A.M., Oliva, G., Garratt, R.C., Souza, D.H.(2008) Toxicon 52: 807-816
- PubMed: 18831982
- DOI: https://doi.org/10.1016/j.toxicon.2008.08.021
- Primary Citation of Related Structures:
3DSL - PubMed Abstract:
Bothropasin is a 48kDa hemorrhagic PIII snake venom metalloprotease (SVMP) isolated from Bothrops jararaca, containing disintegrin/cysteine-rich adhesive domains. Here we present the crystal structure of bothropasin complexed with the inhibitor POL647. The catalytic domain consists of a scaffold of two subdomains organized similarly to those described for other SVMPs, including the zinc and calcium-binding sites. The free cysteine residue Cys189 is located within a hydrophobic core and it is not available for disulfide bonding or other interactions. There is no identifiable secondary structure for the disintegrin domain, but instead it is composed mostly of loops stabilized by seven disulfide bonds and by two calcium ions. The ECD region is in a loop and is structurally related to the RGD region of RGD disintegrins, which are derived from PII SVMPs. The ECD motif is stabilized by the Cys277-Cys310 disulfide bond (between the disintegrin and cysteine-rich domains) and by one calcium ion. The side chain of Glu276 of the ECD motif is exposed to solvent and free to make interactions. In bothropasin, the HVR (hyper-variable region) described for other PIII SVMPs in the cysteine-rich domain, presents a well-conserved sequence with respect to several other PIII members from different species. We propose that this subset be referred to as PIII-HCR (highly conserved region) SVMPs. The differences in the disintegrin-like, cysteine-rich or disintegrin-like cysteine-rich domains may be involved in selecting target binding, which in turn could generate substrate diversity or specificity for the catalytic domain.
- Departamento de Física e Informática, Instituto de Física de São Carlos, USP, São Carlos-SP CEP 13560-970, Brazil.
Organizational Affiliation: