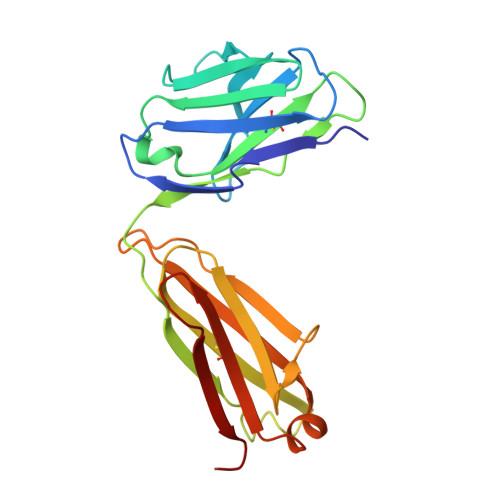
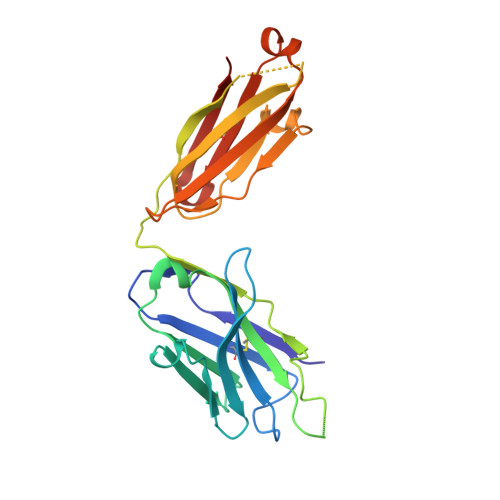
Structural constraints imposed by the conserved fusion peptide on the HIV-1 gp41 epitope recognized by the broadly neutralizing antibody 2F5.
de la Arada, I., Julien, J.P., de la Torre, B.G., Huarte, N., Andreu, D., Pai, E.F., Arrondo, J.L., Nieva, J.L.(2009) J Phys Chem B 113: 13626-13637
- PubMed: 19754136
- DOI: https://doi.org/10.1021/jp905965h
- Primary Citation of Related Structures:
3DRT, 3EGS - PubMed Abstract:
The HIV-1 gp41 epitope recognized by the broadly neutralizing 2F5 antibody has focused much attention as a suitable target in the design of peptide immunogens. Peptides mimicking the linear 2F5 epitope (2F5ep) are however intrinsically disordered, while the structural constraints existing in the cognate gp41 native structure recognized by the antibody are presently unknown. In recent reports, we have shown that core residues of the amino-terminal fusion peptide (FP) increase MAb2F5 affinity. Here, we have inferred the sequence-specific structural constraints imposed by the FP residues on the 2F5 epitope from the comparison of two hybrid peptides: HybK3, which connects through a flexible tether residues derived from 2F5ep and FP sequences, and scrHybK3, combining 2F5ep and an FP sequence with the conserved core scrambled. Circular dichroism, conventional and two-dimensional correlation infrared spectroscopy, and X-ray diffraction studies revealed specific structural features that were dependent on the exact FP sequence, namely, (i) the production with moderate low polarity of an intermediate folded structure enriched in beta-turns and alpha-helix; (ii) the existence in this intermediate of a thermotropic conformational transition taking place at ca. 18-20 degrees C, consistent with the conversion of 3(10)-helices into beta-turn conformers; and (iii) the presence of a C-terminal alpha-helix in crystals of Fab'-peptide complexes. Those features support the existence of native-like tertiary interactions between FP and 2F5 epitope residues, which might be important to recreate when developing an effective AIDS peptide vaccine.
- Biophysics Unit (CSIC-UPV/EHU) and Department of Biochemistry and Molecular Biology, University of the Basque Country, P.O. Box 644, 48080 Bilbao, Spain.
Organizational Affiliation: