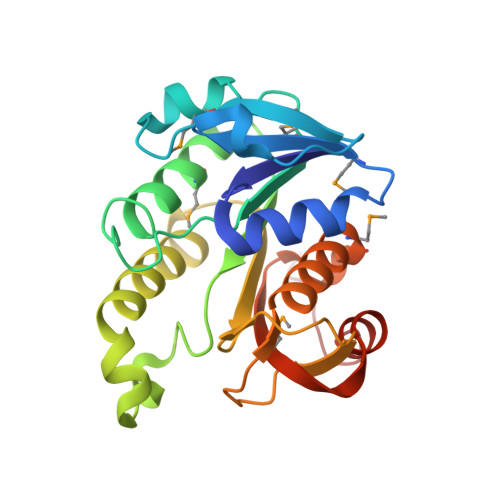
Crystal structure of the oxidoreductase ylbE from Lactococcus lactis, Northeast Structural Genomics Consortium Target KR121.
Forouhar, F., Neely, H., Seetharaman, J., Mao, L., Xiao, R., Ciccosanti, C., Foote, E.L., Lee, D., Everett, J.K., B Acton, T., Montelione, G.T., Tong, L., Hunt, J.F.To be published.