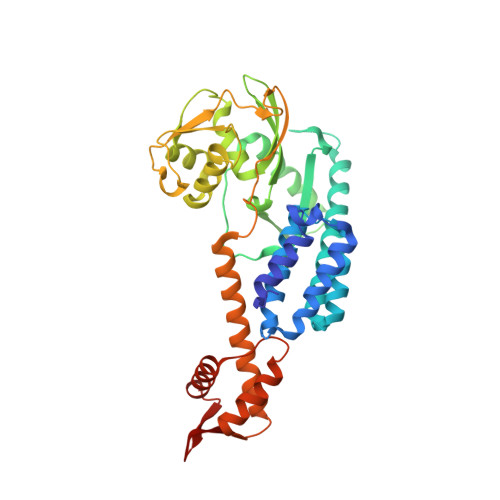
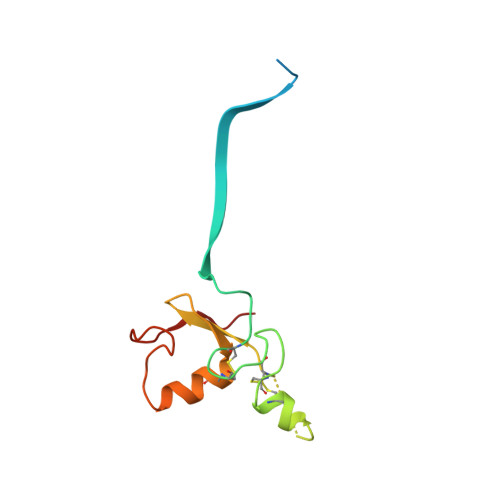
Structural insights into NEDD8 activation of cullin-RING ligases: conformational control of conjugation.
Duda, D.M., Borg, L.A., Scott, D.C., Hunt, H.W., Hammel, M., Schulman, B.A.(2008) Cell 134: 995-1006
- PubMed: 18805092
- DOI: https://doi.org/10.1016/j.cell.2008.07.022
- Primary Citation of Related Structures:
3DPL, 3DQV - PubMed Abstract:
Cullin-RING ligases (CRLs) comprise the largest ubiquitin E3 subclass, in which a central cullin subunit links a substrate-binding adaptor with an E2-binding RING. Covalent attachment of the ubiquitin-like protein NEDD8 to a conserved C-terminal domain (ctd) lysine stimulates CRL ubiquitination activity and prevents binding of the inhibitor CAND1. Here we report striking conformational rearrangements in the crystal structure of NEDD8~Cul5(ctd)-Rbx1 and SAXS analysis of NEDD8~Cul1(ctd)-Rbx1 relative to their unmodified counterparts. In NEDD8ylated CRL structures, the cullin WHB and Rbx1 RING subdomains are dramatically reoriented, eliminating a CAND1-binding site and imparting multiple potential catalytic geometries to an associated E2. Biochemical analyses indicate that the structural malleability is important for both CRL NEDD8ylation and subsequent ubiquitination activities. Thus, our results point to a conformational control of CRL activity, with ligation of NEDD8 shifting equilibria to disfavor inactive CAND1-bound closed architectures, and favor dynamic, open forms that promote polyubiquitination.
- Howard Hughes Medical Institute, St Jude Children's Research Hospital, Memphis, TN 38105, USA.
Organizational Affiliation: