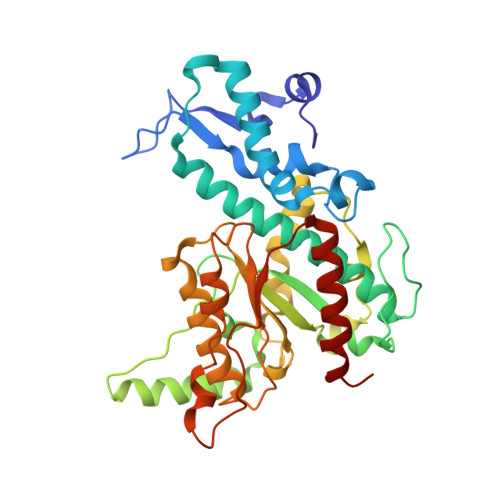
The structure of SgrAI bound to DNA; recognition of an 8 base pair target.
Dunten, P.W., Little, E.J., Gregory, M.T., Manohar, V.M., Dalton, M., Hough, D., Bitinaite, J., Horton, N.C.(2008) Nucleic Acids Res 36: 5405-5416
- PubMed: 18701646
- DOI: https://doi.org/10.1093/nar/gkn510
- Primary Citation of Related Structures:
3DPG, 3DVO, 3DW9 - PubMed Abstract:
The three-dimensional X-ray crystal structure of the 'rare cutting' type II restriction endonuclease SgrAI bound to cognate DNA is presented. SgrAI forms a dimer bound to one duplex of DNA. Two Ca(2+) bind in the enzyme active site, with one ion at the interface between the protein and DNA, and the second bound distal from the DNA. These sites are differentially occupied by Mn(2+), with strong binding at the protein-DNA interface, but only partial occupancy of the distal site. The DNA remains uncleaved in the structures from crystals grown in the presence of either divalent cation. The structure of the dimer of SgrAI is similar to those of Cfr10I, Bse634I and NgoMIV, however no tetrameric structure of SgrAI is observed. DNA contacts to the central CCGG base pairs of the SgrAI canonical target sequence (CR|CCGGYG, | marks the site of cleavage) are found to be very similar to those in the NgoMIV/DNA structure (target sequence G|CCGGC). Specificity at the degenerate YR base pairs of the SgrAI sequence may occur via indirect readout using DNA distortion. Recognition of the outer GC base pairs occurs through a single contact to the G from an arginine side chain located in a region unique to SgrAI.
- Stanford Synchrotron Radiation Laboratory, Stanford University, Menlo Park, CA 94025, USA.
Organizational Affiliation: