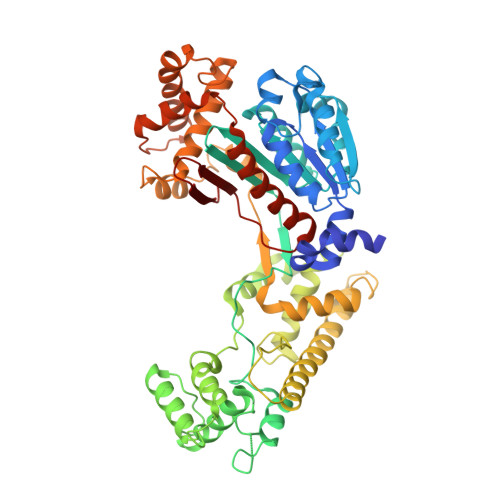
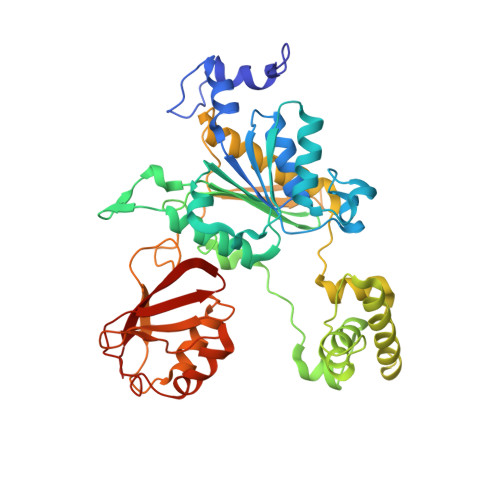
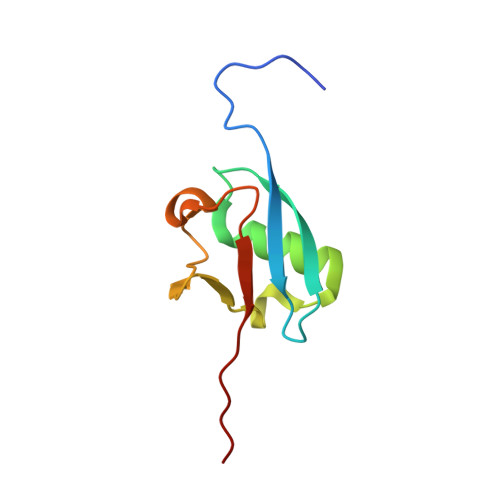
Structural dissection of a gating mechanism preventing misactivation of ubiquitin by NEDD8's E1.
Souphron, J., Waddell, M.B., Paydar, A., Tokgoz-Gromley, Z., Roussel, M.F., Schulman, B.A.(2008) Biochemistry 47: 8961-8969
- PubMed: 18652489
- DOI: https://doi.org/10.1021/bi800604c
- Primary Citation of Related Structures:
3DBH, 3DBL, 3DBR - PubMed Abstract:
Post-translational covalent modification by ubiquitin and ubiquitin-like proteins (UBLs) is a major eukaryotic mechanism for regulating protein function. In general, each UBL has its own E1 that serves as the entry point for a cascade. The E1 first binds the UBL and catalyzes adenylation of the UBL's C-terminus, prior to promoting UBL transfer to a downstream E2. Ubiquitin's Arg 72, which corresponds to Ala72 in the UBL NEDD8, is a key E1 selectivity determinant: swapping ubiquitin and NEDD8 residue 72 identity was shown previously to swap their E1 specificity. Correspondingly, Arg190 in the UBA3 subunit of NEDD8's heterodimeric E1 (the APPBP1-UBA3 complex), which corresponds to a Gln in ubiquitin's E1 UBA1, is a key UBL selectivity determinant. Here, we dissect this specificity with biochemical and X-ray crystallographic analysis of APPBP1-UBA3-NEDD8 complexes in which NEDD8's residue 72 and UBA3's residue 190 are substituted with different combinations of Ala, Arg, or Gln. APPBP1-UBA3's preference for NEDD8's Ala72 appears to be indirect, due to proper positioning of UBA3's Arg190. By contrast, our data are consistent with direct positive interactions between ubiquitin's Arg72 and an E1's Gln. However, APPBP1-UBA3's failure to interact with a UBL having Arg72 is not due to a lack of this favorable interaction, but rather arises from UBA3's Arg190 acting as a negative gate. Thus, parallel residues from different UBL pathways can utilize distinct mechanisms to dictate interaction selectivity, and specificity can be amplified by barriers that prevent binding to components of different conjugation cascades.
- Department of Structural Biology, St. Jude Children's Research Hospital, Memphis, Tennessee 38105, USA.
Organizational Affiliation: