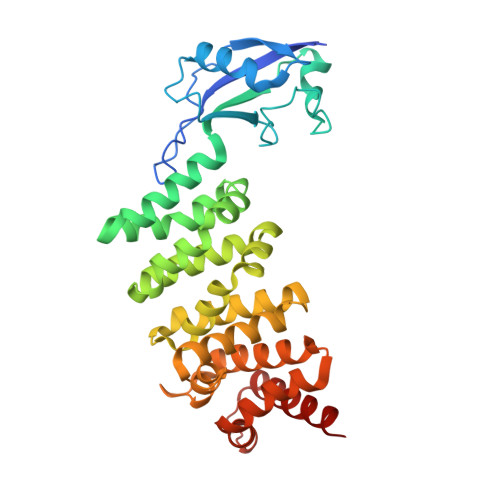
The Human Formin FHOD1 Contains a Bipartite Structure of FH3 and GTPase-Binding Domains Required for Activation.
Schulte, A., Stolp, B., Schonichen, A., Pylypenko, O., Rak, A., Fackler, O.T., Geyer, M.(2008) Structure 16: 1313-1323
- PubMed: 18786395
- DOI: https://doi.org/10.1016/j.str.2008.06.008
- Primary Citation of Related Structures:
3DAD - PubMed Abstract:
Formins induce the nucleation and polymerization of unbranched actin filaments. They share three homology domains required for profilin binding, actin polymerization, and regulation. Diaphanous-related formins (DRFs) are activated by GTPases of the Rho/Rac family, whose interaction with the N-terminal formin domain is thought to displace a C-terminal Diaphanous-autoregulatory domain (DAD). We have determined the structure of the N-terminal domains of FHOD1 consisting of a GTPase-binding domain (GBD) and the DAD-recognition domain FH3. In contrast to the formin mDia1, the FHOD1-GBD reveals a ubiquitin superfold as found similarly in c-Raf1 or PI3 kinase. This GBD is recruited by Rac and Ras GTPases in cells and plays an essential role for FHOD1-mediated actin remodeling. The FHOD1-FH3 domain is composed of five armadillo repeats, similarly to other formins. Mutation of one residue in the predicted DAD-interaction surface efficiently activates FHOD1 in cells. These results demonstrate that DRFs have evolved different molecular solutions to govern their autoregulation and GTPase specificity.
- Abteilung Physikalische Biochemie, Max-Planck-Institut für Molekulare Physiologie, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany.
Organizational Affiliation: