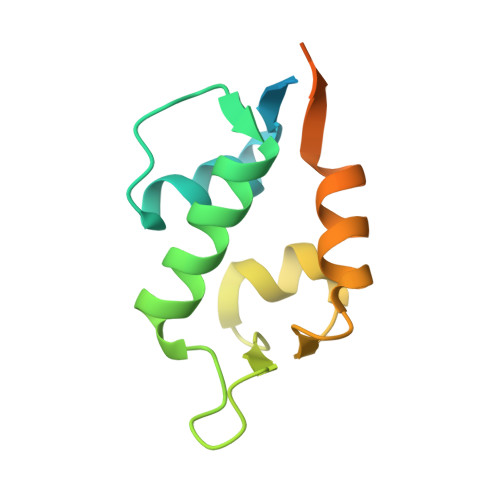
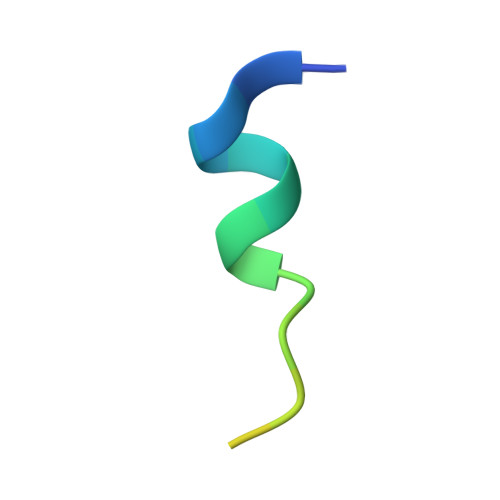
Structure of the human Mdmx protein bound to the p53 tumor suppressor transactivation domain.
Popowicz, G.M., Czarna, A., Holak, T.A.(2008) Cell Cycle 7: 2441-2443
- PubMed: 18677113
- DOI: https://doi.org/10.4161/cc.6365
- Primary Citation of Related Structures:
3DAB, 3DAC - PubMed Abstract:
The Mdmx oncoprotein has only recently emerged as a critical-independent to Mdm2-regulator of p53 activation. We have determined the crystal structure of the N-terminal domain of human Mdmx bound to a 15-residue transactivation domain peptide of human p53. The structure shows why antagonists of the Mdm2 binding to p53 are ineffective in the Mdmx-p53 interaction.