There is a baby in the bath water: AcrB contamination is a major problem in membrane-protein crystallization.
Veesler, D., Blangy, S., Cambillau, C., Sciara, G.(2008) Acta Crystallogr Sect F Struct Biol Cryst Commun 64: 880-885
- PubMed: 18931428
- DOI: https://doi.org/10.1107/S1744309108028248
- Primary Citation of Related Structures:
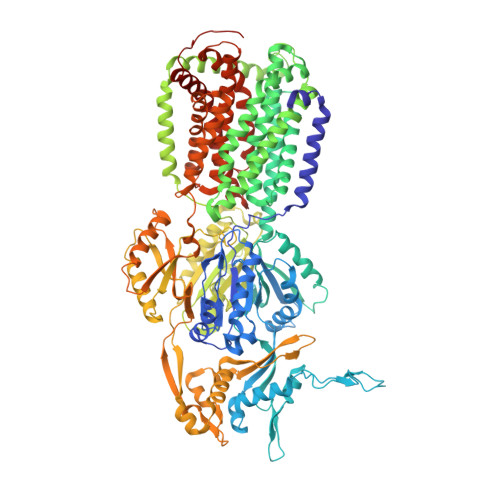
3D9B - PubMed Abstract:
In the course of a crystallographic study of the Methanosarcina mazei CorA transporter, the membrane protein was obtained with at least 95% purity and was submitted to crystallization trials. Small crystals (<100 microm) were grown that diffracted to 3.42 A resolution and belonged to space group R32, with unit-cell parameters a = b = 145.74, c = 514.0 A. After molecular-replacement attempts using available CorA structures as search models failed to yield a solution, it was discovered that the crystals consisted of an Escherichia coli contaminating protein, acriflavine resistance protein B (AcrB), that was present at less than 5% in the protein preparations. AcrB contamination is a major problem when expressing membrane proteins in E. coli since it binds naturally to immobilized metal-ion affinity chromatography (IMAC) resins. Here, the structure is compared with previously deposited AcrB structures and strategies are proposed to avoid this contamination.
- Architecture et Fonction des Macromolécules Biologiques, CNRS et Universités d'Aix-Marseille I et II, UMR 6098, Case 932, 163 Avenue de Luminy, 13288 Marseille CEDEX 9, France. david.veesler@afmb.univ-mrs.fr
Organizational Affiliation: