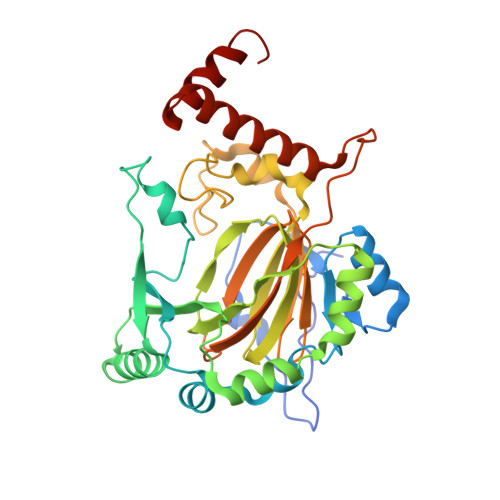
Evidence that two enzyme-derived histidine ligands are sufficient for iron binding and catalysis by factor inhibiting HIF (FIH).
Hewitson, K.S., Holmes, S.L., Ehrismann, D., Hardy, A.P., Chowdhury, R., Schofield, C.J., McDonough, M.A.(2008) J Biological Chem 283: 25971-25978
- PubMed: 18611856
- DOI: https://doi.org/10.1074/jbc.M804999200
- Primary Citation of Related Structures:
2ILM, 3D8C - PubMed Abstract:
A 2-His-1-carboxylate triad of iron binding residues is present in many non-heme iron oxygenases including the Fe(II) and 2-oxoglutarate (2OG)-dependent dioxygenases. Three variants (D201A, D201E, and D201G) of the iron binding Asp-201 residue of an asparaginyl hydroxylase, factor inhibiting HIF (FIH), were made and analyzed. FIH-D201A and FIH-D201E did not catalyze asparaginyl hydroxylation, but in the presence of a reducing agent, they displayed enhanced 2OG turnover when compared with wild-type FIH. Turnover of 2OG by FIH-D201A was significantly stimulated by the addition of HIF-1alpha(786-826) peptide. Like FIH-D201A and D201E, the D201G variant enhanced 2OG turnover but rather unexpectedly catalyzed asparaginyl hydroxylation. Crystal structures of the FIH-D201A and D201G variants in complex with Fe(II)/Zn(II), 2OG, and HIF-1alpha(786-826/788-806) implied that only two FIH-based residues (His-199 and His-279) are required for metal binding. The results indicate that variation of 2OG-dependent dioxygenase iron-ligating residues as a means of functional assignment should be treated with caution. The results are of mechanistic interest in the light of recent biochemical and structural analyses of non-heme iron and 2OG-dependent halogenases that are similar to the FIH-D201A/G variants in that they use only two His-residues to ligate iron.
- Chemistry Research Laboratory, The Department of Chemistry, University of Oxford, Mansfield Road, Oxford OX1 3TA, United Kingdom.
Organizational Affiliation: