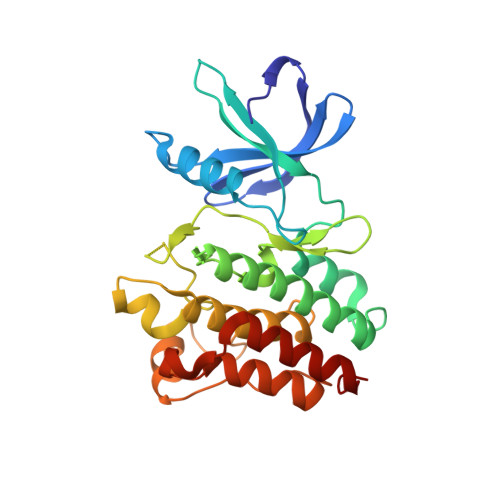
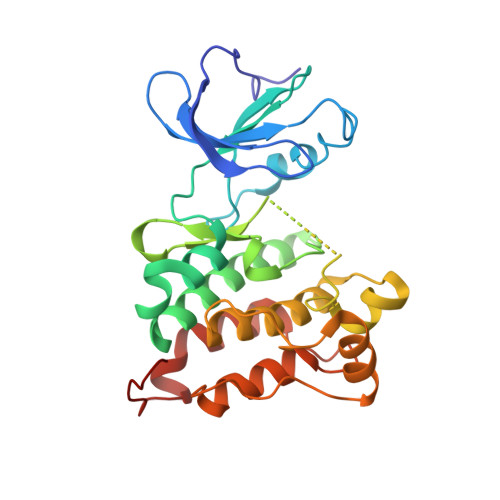
Structural basis for the recognition of c-Src by its inactivator Csk.
Levinson, N.M., Seeliger, M.A., Cole, P.A., Kuriyan, J.(2008) Cell 134: 124-134
- PubMed: 18614016
- DOI: https://doi.org/10.1016/j.cell.2008.05.051
- Primary Citation of Related Structures:
3D7T, 3D7U - PubMed Abstract:
The catalytic activity of the Src family of tyrosine kinases is suppressed by phosphorylation on a tyrosine residue located near the C terminus (Tyr 527 in c-Src), which is catalyzed by C-terminal Src Kinase (Csk). Given the promiscuity of most tyrosine kinases, it is remarkable that the C-terminal tails of the Src family kinases are the only known targets of Csk. We have determined the crystal structure of a complex between the kinase domains of Csk and c-Src at 2.9 A resolution, revealing that interactions between these kinases position the C-terminal tail of c-Src at the edge of the active site of Csk. Csk cannot phosphorylate substrates that lack this docking mechanism because the conventional substrate binding site used by most tyrosine kinases to recognize substrates is destabilized in Csk by a deletion in the activation loop.
- Department of Molecular and Cell Biology, Department of Chemistry, Howard Hughes Medical Institute, California Institute for Quantitative Biosciences (QB3), University of California, Berkeley, Berkeley, CA 94720, USA.
Organizational Affiliation: