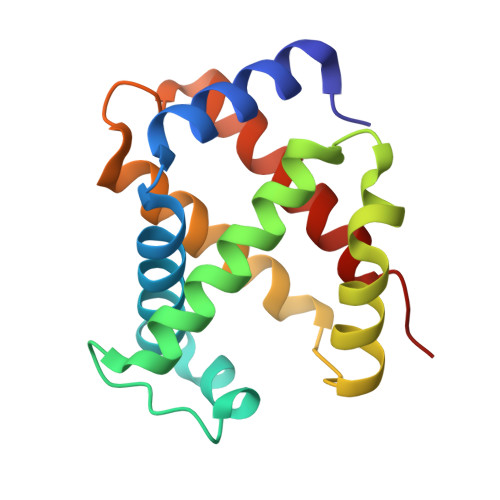
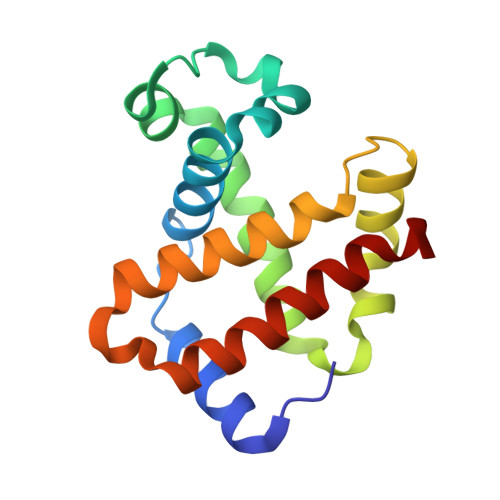
The nitrite anion binds to human hemoglobin via the uncommon O-nitrito mode.
Yi, J., Safo, M.K., Richter-Addo, G.B.(2008) Biochemistry 47: 8247-8249
- PubMed: 18630930
- DOI: https://doi.org/10.1021/bi801015c
- Primary Citation of Related Structures:
3D7O - PubMed Abstract:
The nitrite anion is known to oxidize and degrade hemoglobin (Hb). Recent literature reports suggest a nitrite reductase activity for Hb, converting nitrite into nitric oxide. Surprisingly, no structural information about Hb-nitrite interactions has been reported. We have determined the crystal structure of the ferric Hb-nitrite complex at 1.80 A resolution. The nitrite ligand adopts the uncommon O-nitrito binding mode. In addition, the nitrito conformations in the alpha and beta subunits are different, reflecting subtle effects of the distal His in orienting the nitrite ligand in the O-nitrito binding mode.
- Department of Chemistry and Biochemistry, University of Oklahoma, Norman, Oklahoma 73019, USA.
Organizational Affiliation: