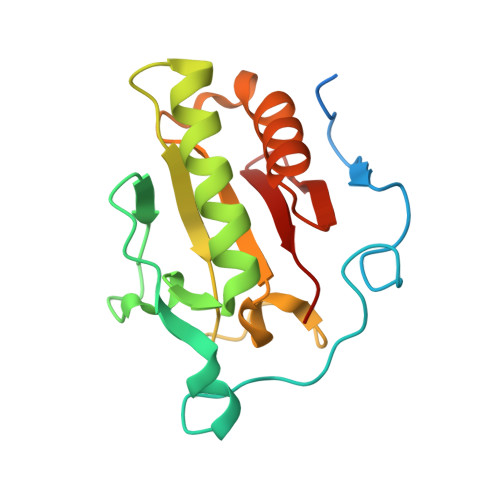
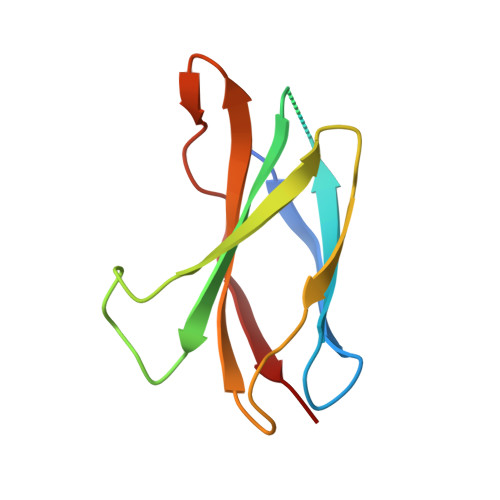
The mode of Hedgehog binding to Ihog homologues is not conserved across different phyla.
McLellan, J.S., Zheng, X., Hauk, G., Ghirlando, R., Beachy, P.A., Leahy, D.J.(2008) Nature 455: 979-983
- PubMed: 18794898
- DOI: https://doi.org/10.1038/nature07358
- Primary Citation of Related Structures:
3D1M - PubMed Abstract:
Hedgehog (Hh) proteins specify tissue pattern in metazoan embryos by forming gradients that emanate from discrete sites of expression and elicit concentration-dependent cellular differentiation or proliferation responses. Cellular responses to Hh and the movement of Hh through tissues are both precisely regulated, and abnormal Hh signalling has been implicated in human birth defects and cancer. Hh signalling is mediated by its amino-terminal domain (HhN), which is dually lipidated and secreted as part of a multivalent lipoprotein particle. Reception of the HhN signal is modulated by several cell-surface proteins on responding cells, including Patched (Ptc), Smoothened (Smo), Ihog (known as CDO or CDON in mammals) and the vertebrate-specific proteins Hip (also known as Hhip) and Gas1 (ref. 11). Drosophila Ihog and its vertebrate homologues CDO and BOC contain multiple immunoglobulin and fibronectin type III (FNIII) repeats, and the first FNIII repeat of Ihog binds Drosophila HhN in a heparin-dependent manner. Surprisingly, pull-down experiments suggest that a mammalian Sonic hedgehog N-terminal domain (ShhN) binds a non-orthologous FNIII repeat of CDO. Here we report biochemical, biophysical and X-ray structural studies of a complex between ShhN and the third FNIII repeat of CDO. We show that the ShhN-CDO interaction is completely unlike the HhN-Ihog interaction and requires calcium, which binds at a previously undetected site on ShhN. This site is conserved in nearly all Hh proteins and is a hotspot for mediating interactions between ShhN and CDO, Ptc, Hip and Gas1. Mutations in vertebrate Hh proteins causing holoprosencephaly and brachydactyly type A1 map to this calcium-binding site and disrupt interactions with these partners.
- Department of Biophysics and Biophysical Chemistry, Johns Hopkins University School of Medicine, Baltimore, Maryland 21205, USA.
Organizational Affiliation: