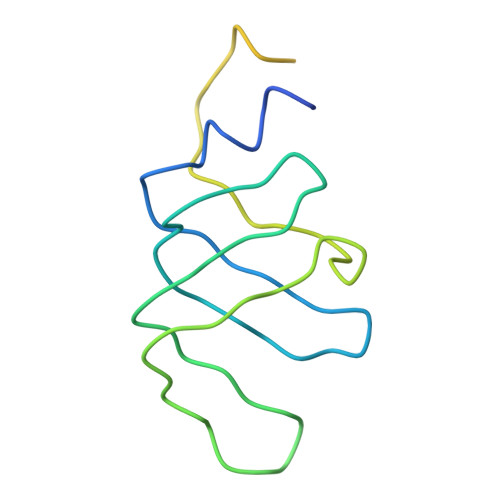
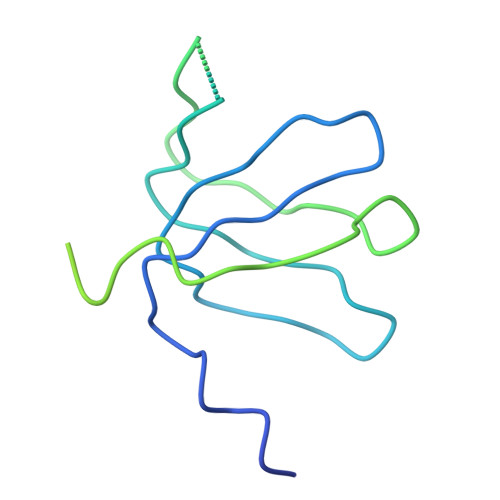
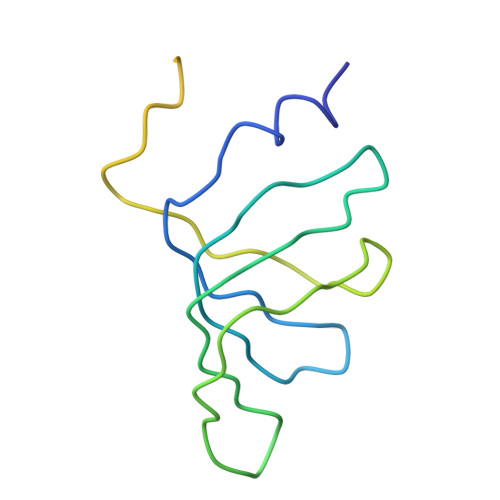
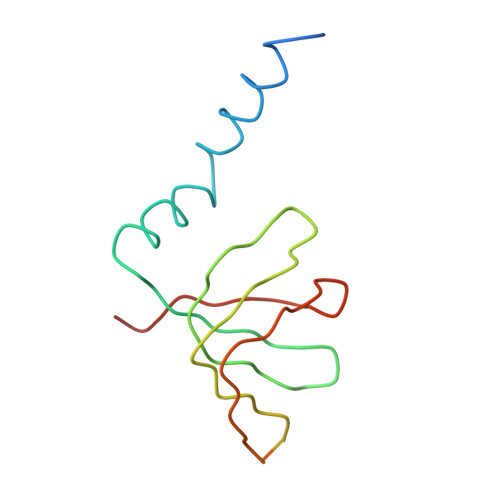
Crystal structure of human spliceosomal U1 snRNP at 5.5 A resolution.
Pomeranz Krummel, D.A., Oubridge, C., Leung, A.K., Li, J., Nagai, K.(2009) Nature 458: 475-480
- PubMed: 19325628
- DOI: https://doi.org/10.1038/nature07851
- Primary Citation of Related Structures:
3CW1 - PubMed Abstract:
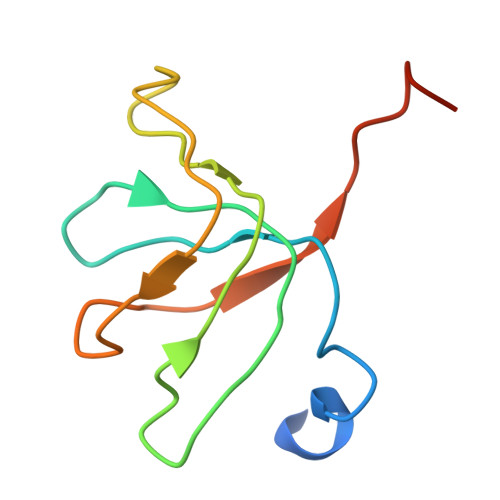
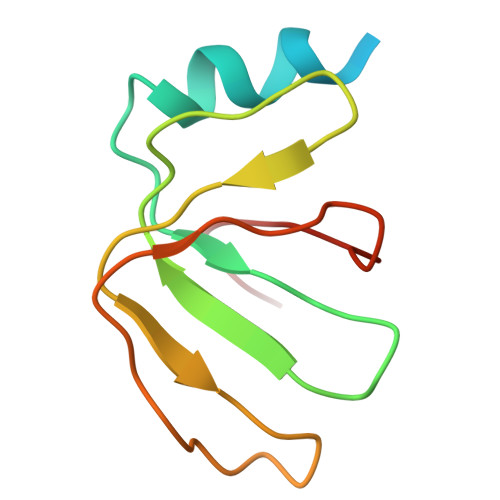
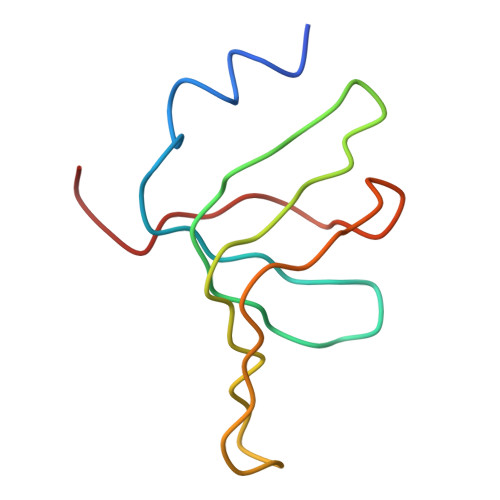
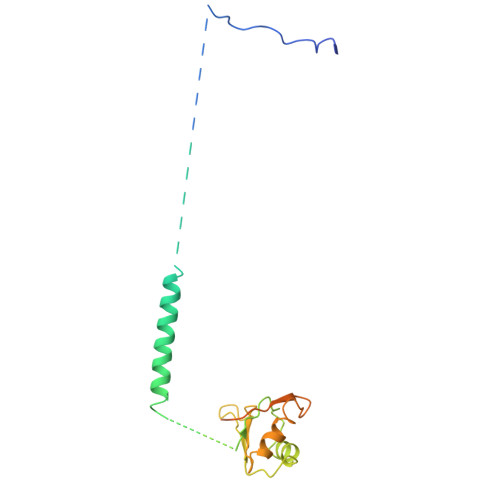
Human spliceosomal U1 small nuclear ribonucleoprotein particles (snRNPs), which consist of U1 small nuclear RNA and ten proteins, recognize the 5' splice site within precursor messenger RNAs and initiate the assembly of the spliceosome for intron excision. An electron density map of the functional core of U1 snRNP at 5.5 A resolution has enabled us to build the RNA and, in conjunction with site-specific labelling of individual proteins, to place the seven Sm proteins, U1-C and U1-70K into the map. Here we present the detailed structure of a spliceosomal snRNP, revealing a hierarchical network of intricate interactions between subunits. A striking feature is the amino (N)-terminal polypeptide of U1-70K, which extends over a distance of 180 A from its RNA binding domain, wraps around the core domain consisting of the seven Sm proteins and finally contacts U1-C, which is crucial for 5'-splice-site recognition. The structure of U1 snRNP provides insights into U1 snRNP assembly and suggests a possible mechanism of 5'-splice-site recognition.
- MRC Laboratory of Molecular Biology, Hills Road, Cambridge CB2 0QH, UK.
Organizational Affiliation: