Crystal structure of the MHC class II molecule I-Ag7 in complex with the peptide GAD221-235
Yoshida, K., Corper, A.L., Puckett, C., Sim, J., Valerie, M.-D., Jabri, B., Wilson, I.A., Teyton, L.To be published.
Experimental Data Snapshot
Starting Model: experimental
View more details
Entity ID: 1 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| H-2 class II histocompatibility antigen, A-D alpha chain | 190 | Mus musculus | Mutation(s): 0 Gene Names: H2-Aa | 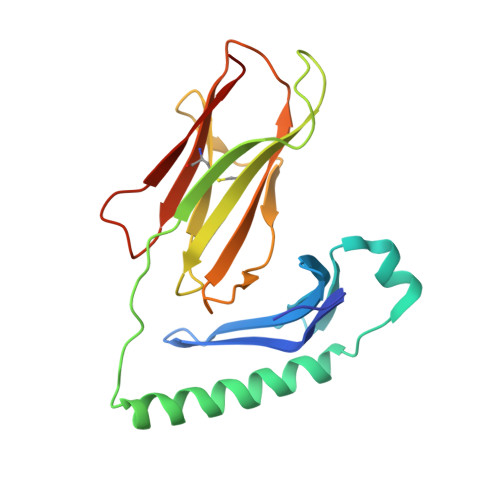 | |
UniProt | |||||
Find proteins for P04228 (Mus musculus) Explore P04228 Go to UniProtKB: P04228 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Group | P04228 | ||||
Glycosylation | |||||
| Glycosylation Sites: 1 | |||||
Sequence AnnotationsExpand | |||||
| |||||
Entity ID: 2 | |||||
|---|---|---|---|---|---|
| Molecule | Chains | Sequence Length | Organism | Details | Image |
| MHC class II H2-IA-beta chain linked to GAD221-235 peptide | 222 | Mus musculus | Mutation(s): 0 EC: 4.1.1.15 | 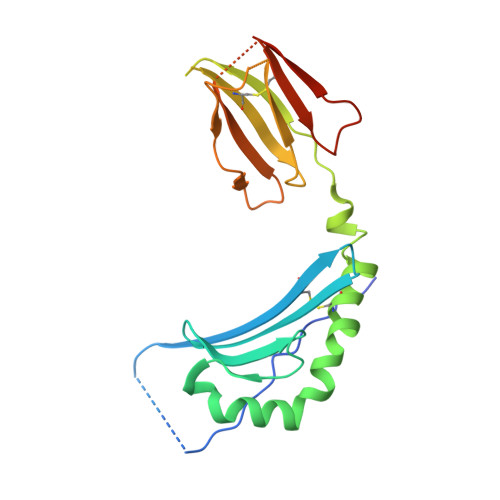 | |
UniProt | |||||
Find proteins for Q05683 (Rattus norvegicus) Explore Q05683 Go to UniProtKB: Q05683 | |||||
Find proteins for Q31135 (Mus musculus) Explore Q31135 Go to UniProtKB: Q31135 | |||||
Entity Groups | |||||
| Sequence Clusters | 30% Identity50% Identity70% Identity90% Identity95% Identity100% Identity | ||||
| UniProt Groups | Q31135Q05683 | ||||
Sequence AnnotationsExpand | |||||
| |||||
| Ligands 2 Unique | |||||
|---|---|---|---|---|---|
| ID | Chains | Name / Formula / InChI Key | 2D Diagram | 3D Interactions | |
| EPE Query on EPE | D [auth A] | 4-(2-HYDROXYETHYL)-1-PIPERAZINE ETHANESULFONIC ACID C8 H18 N2 O4 S JKMHFZQWWAIEOD-UHFFFAOYSA-N |  | ||
| NAG Query on NAG | C [auth A] | 2-acetamido-2-deoxy-beta-D-glucopyranose C8 H15 N O6 OVRNDRQMDRJTHS-FMDGEEDCSA-N |  | ||
| Length ( Å ) | Angle ( ˚ ) |
|---|---|
| a = 55.832 | α = 90 |
| b = 55.832 | β = 90 |
| c = 338.834 | γ = 90 |
| Software Name | Purpose |
|---|---|
| PHASER | phasing |
| PHENIX | refinement |
| PDB_EXTRACT | data extraction |
| ADSC | data collection |