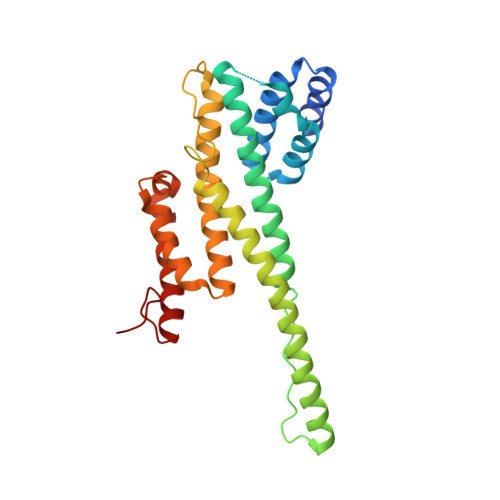
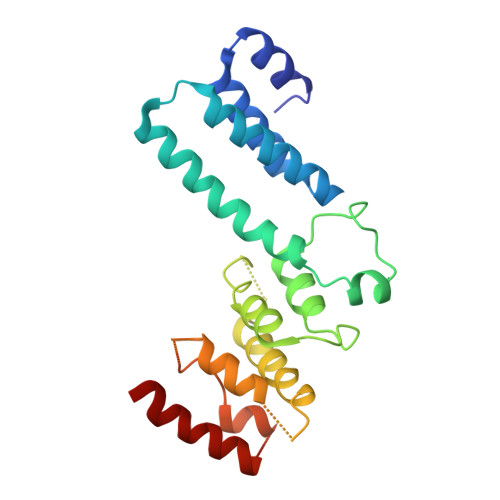
Structural and functional studies of Nup107/Nup133 interaction and its implications for the architecture of the nuclear pore complex.
Boehmer, T., Jeudy, S., Berke, I.C., Schwartz, T.U.(2008) Mol Cell 30: 721-731
- PubMed: 18570875
- DOI: https://doi.org/10.1016/j.molcel.2008.04.022
- Primary Citation of Related Structures:
3CQC, 3CQG - PubMed Abstract:
Nuclear pore complexes (NPCs) are 40-60 MDa protein assemblies embedded in the nuclear envelope of eukaryotic cells. NPCs exclusively mediate all transport between cytoplasm and nucleus. The nucleoporins that build the NPC are arranged in a stable core of module-like subcomplexes with eight-fold rotational symmetry. To gain insight into the intricate assembly of the NPC, we have solved the crystal structure of a protein complex between two nucleoporins, human Nup107 and Nup133. Both proteins form elongated structures that interact tightly via a compact interface in tail-to-tail fashion. Additional experiments using structure-guided mutants show that Nup107 is the critical anchor for Nup133 to the NPC, positioning Nup133 at the periphery of the NPC. The significant topological differences between Nup107 and Nup133 suggest that *-helical nucleoporin domains of the NPC scaffold fall in different classes and fulfill largely nonredundant functions.
- Department of Biology, Massachusetts Institute of Technology, 77 Massachusetts Avenue, Cambridge, MA 02139, USA.
Organizational Affiliation: