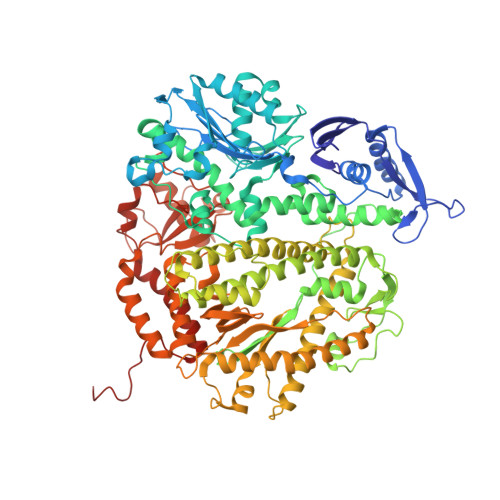
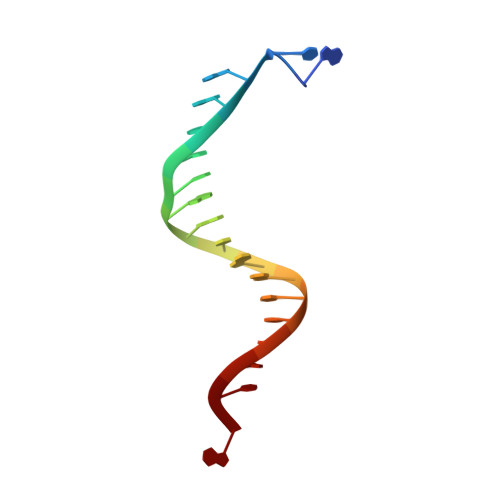
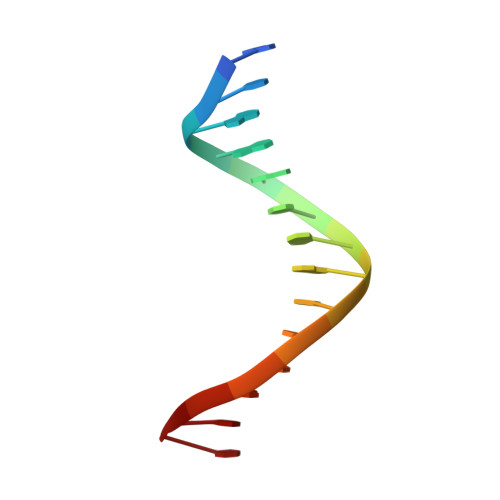
Characterization of a replicative DNA polymerase mutant with reduced fidelity and increased translesion synthesis capacity.
Zhong, X., Pedersen, L.C., Kunkel, T.A.(2008) Nucleic Acids Res 36: 3892-3904
- PubMed: 18503083
- DOI: https://doi.org/10.1093/nar/gkn312
- Primary Citation of Related Structures:
3CQ8 - PubMed Abstract:
Changing a highly conserved amino acid in motif A of any of the four yeast family B DNA polymerases, DNA polymerase alpha, delta, epsilon or zeta, results in yeast strains with elevated mutation rates. In order to better understand this phenotype, we have performed structure-function studies of homologous mutants of RB69 DNA polymerase (RB69 pol), a structural model for family B members. When Leu415 in RB69 pol is replaced with phenylalanine or glycine, the mutant polymerases retain high-catalytic efficiency for correct nucleotide incorporation, yet have increased error rates due to increased misinsertion, increased mismatch extension and inefficient proofreading. The Leu415Phe mutant also has increased dNTP insertion efficiency opposite a template 8-oxoG and opposite an abasic site. The 2.5 A crystal structure of a ternary complex of RB69 L415F pol with a correctly base-paired incoming dTTP reveals that the phenylalanine ring is accommodated within a cavity seen in the wild-type enzyme, without steric clash or major change in active site geometry, consistent with retention of high-catalytic efficiency for correct incorporation. In addition, slight structural differences were observed that could be relevant to the reduced fidelity of L415F RB69 pol.
- Laboratory of Molecular Genetics and Laboratory of Structural Biology National Institute of Environmental Health Sciences, NIH, DHHS Research Triangle Park, NC 27709, USA.
Organizational Affiliation: