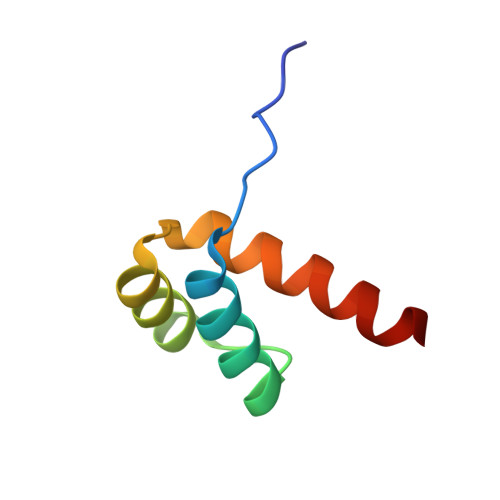
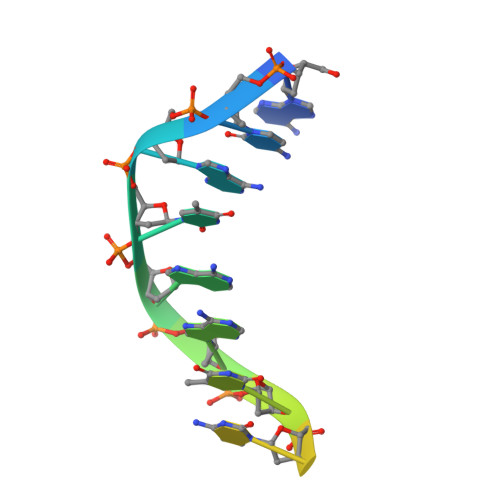
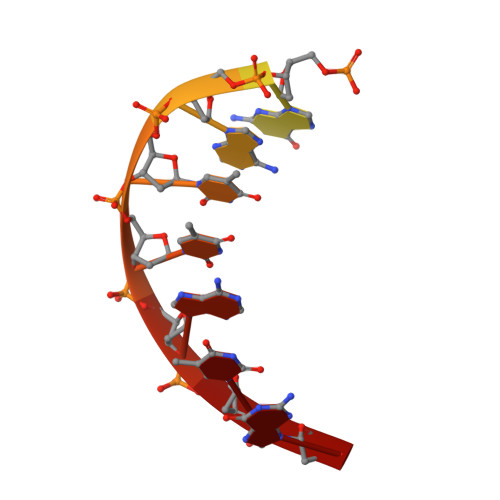
Structural Basis for DNA Recognition by the Human PAX3 Homeodomain.
Birrane, G., Soni, A., Ladias, J.A.(2009) Biochemistry 48: 1148-1155
- PubMed: 19199574
- DOI: https://doi.org/10.1021/bi802052y
- Primary Citation of Related Structures:
3CMY - PubMed Abstract:
The transcription regulatory protein PAX3 binds to cognate DNA sequences through two DNA-binding domains, a paired domain and a homeodomain, and has important functions during neurogenesis and myogenesis. In humans, mutations in the PAX3 gene cause Waardenburg syndrome, whereas a chromosomal translocation that generates a PAX3-FOXO1 fusion gene is associated with the development of alveolar rhabdomyosarcoma. We have determined the crystal structure of the human PAX3 homeodomain in complex with a palindromic DNA containing two inverted TAATC sequences at 1.95 A resolution. Two homeodomains bind to DNA as a symmetric dimer, inducing a 3 degrees bend in the DNA helix. The N-terminal arm of the homeodomain inserts into the minor groove and makes direct and water-mediated interactions with bases and the sugar-phosphate backbone. The recognition helix fits directly into the major groove, and an elaborate network of structurally conserved water molecules mediates the majority of protein-DNA interactions. The structure elucidates the role of serine 50 in selection of the CG sequence immediately 3' of the TAAT motif by PAX class homeodomains and provides insights into the molecular mechanisms by which certain Waardenburg syndrome-associated missense mutations could destabilize the fold of the PAX3 homeodomain whereas others could affect its interaction with DNA.
- Molecular Medicine Laboratory and Macromolecular Crystallography Unit, Division of Experimental Medicine, Harvard Medical School, Boston, Massachusetts 02115, USA.
Organizational Affiliation: