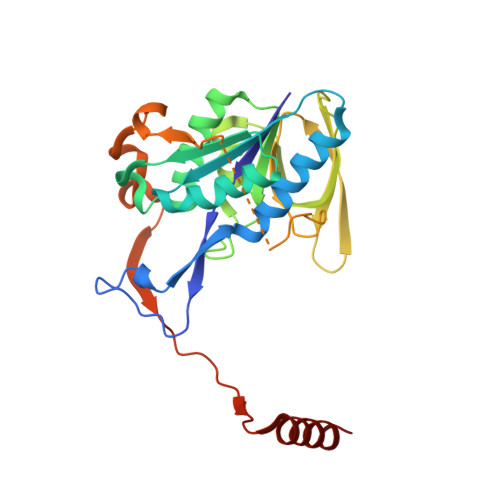
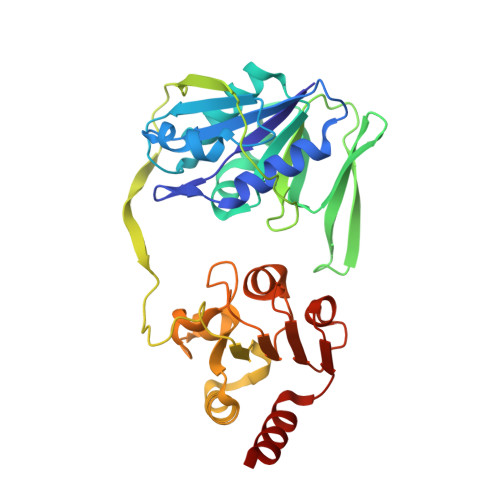
Probing the dynamic interface between trimethylamine dehydrogenase (TMADH) and electron transferring flavoprotein (ETF) in the TMADH-2ETF complex: role of the Arg-alpha237 (ETF) and Tyr-442 (TMADH) residue pair.
Burgess, S.G., Messiha, H.L., Katona, G., Rigby, S.E., Leys, D., Scrutton, N.S.(2008) Biochemistry 47: 5168-5181
- PubMed: 18407658
- DOI: https://doi.org/10.1021/bi800127d
- Primary Citation of Related Structures:
3CLR, 3CLS, 3CLT, 3CLU - PubMed Abstract:
We have used multiple solution state techniques and crystallographic analysis to investigate the importance of a putative transient interaction formed between Arg-alpha237 in electron transferring flavoprotein (ETF) and Tyr-442 in trimethylamine dehydrogenase (TMADH) in complex assembly, electron transfer, and structural imprinting of ETF by TMADH. We have isolated four mutant forms of ETF altered in the identity of the residue at position 237 (alphaR237A, alphaR237K, alphaR237C, and alphaR237E) and with each form studied electron transfer from TMADH to ETF, investigated the reduction potentials of the bound ETF cofactor, and analyzed complex formation. We show that mutation of Arg-alpha237 substantially destabilizes the semiquinone couple of the bound FAD and impedes electron transfer from TMADH to ETF. Crystallographic structures of the mutant ETF proteins indicate that mutation does not perturb the overall structure of ETF, but leads to disruption of an electrostatic network at an ETF domain boundary that likely affects the dynamic properties of ETF in the crystal and in solution. We show that Arg-alpha237 is required for TMADH to structurally imprint the as-purified semiquinone form of wild-type ETF and that the ability of TMADH to facilitate this structural reorganization is lost following (i) redox cycling of ETF, or simple conversion to the oxidized form, and (ii) mutagenesis of Arg-alpha237. We discuss this result in light of recent apparent conflict in the literature relating to the structural imprinting of wild-type ETF. Our studies support a mechanism of electron transfer by conformational sampling as advanced from our previous analysis of the crystal structure of the TMADH-2ETF complex [Leys, D. , Basran, J. , Sutcliffe, M. J., and Scrutton, N. S. (2003) Nature Struct. Biol. 10, 219-225] and point to a key role for the Tyr-442 (TMADH) and Arg-alpha237 (ETF) residue pair in transiently stabilizing productive electron transfer configurations. Our work also points to the importance of Arg-alpha237 in controlling the thermodynamics of electron transfer, the dynamics of ETF, and the protection of reducing equivalents following disassembly of the TMADH-2ETF complex.
- Department of Biochemistry, University of Leicester, Leicester LE1 9HN, UK.
Organizational Affiliation: