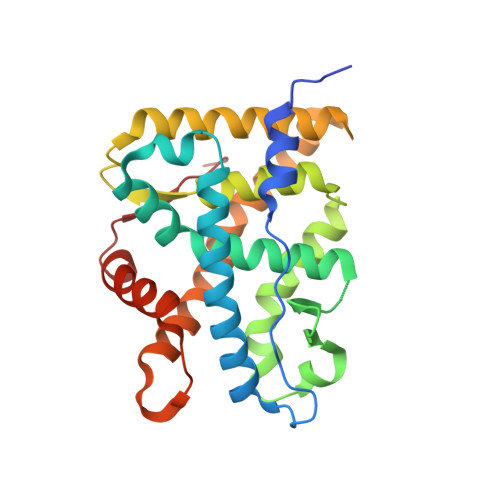
X-ray crystal structure of the novel enhanced-affinity glucocorticoid agonist fluticasone furoate in the glucocorticoid receptor-ligand binding domain.
Biggadike, K., Bledsoe, R.K., Hassell, A.M., Kirk, B.E., McLay, I.M., Shewchuk, L.M., Stewart, E.L.(2008) J Med Chem 51: 3349-3352
- PubMed: 18522385
- DOI: https://doi.org/10.1021/jm800279t
- Primary Citation of Related Structures:
3CLD - PubMed Abstract:
An X-ray crystal structure is reported for the novel enhanced-affinity glucocorticoid agonist fluticasone furoate (FF) in the ligand binding domain of the glucocorticoid receptor. Comparison of this structure with those of dexamethasone and fluticasone propionate shows the 17 alpha furoate ester to occupy more fully the lipophilic 17 alpha pocket on the receptor, which may account for the enhanced glucocorticoid receptor binding of FF.
- kb0903@gsk.com
Organizational Affiliation: