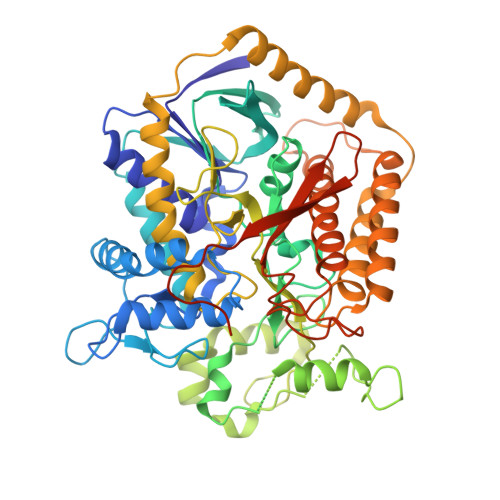
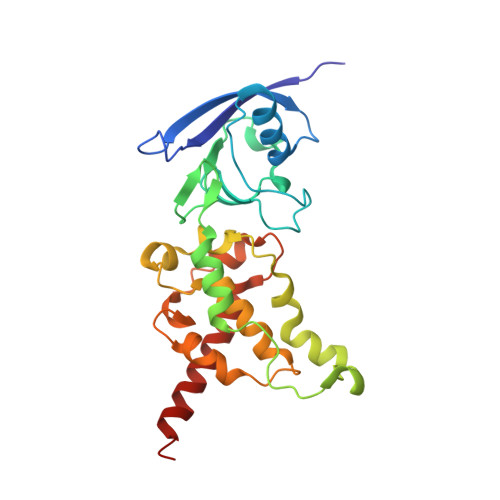
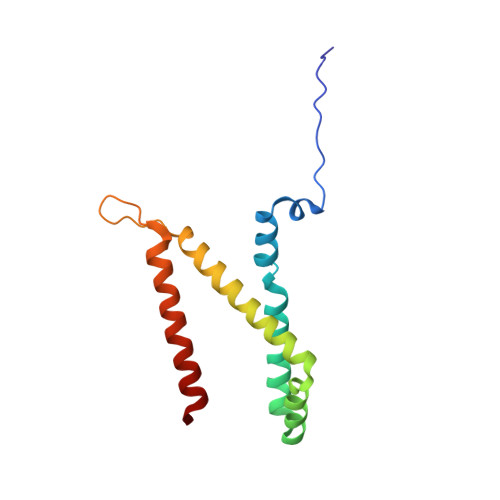
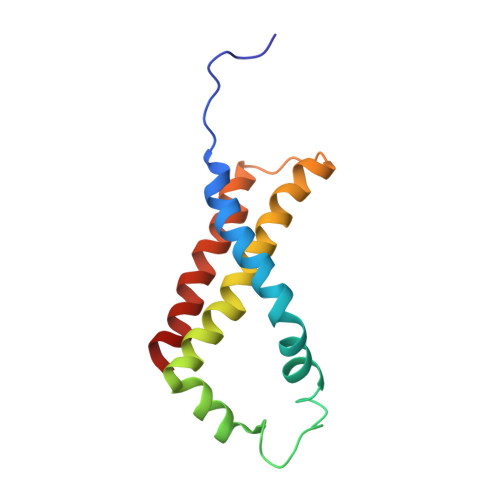
A threonine on the active site loop controls transition state formation in Escherichia coli respiratory complex II.
Tomasiak, T.M., Maklashina, E., Cecchini, G., Iverson, T.M.(2008) J Biological Chem 283: 15460-15468
- PubMed: 18385138
- DOI: https://doi.org/10.1074/jbc.M801372200
- Primary Citation of Related Structures:
3CIR - PubMed Abstract:
In Escherichia coli, the complex II superfamily members succinate:ubiquinone oxidoreductase (SQR) and quinol:fumarate reductase (QFR) participate in aerobic and anaerobic respiration, respectively. Complex II enzymes catalyze succinate and fumarate interconversion at the interface of two domains of the soluble flavoprotein subunit, the FAD binding domain and the capping domain. An 11-amino acid loop in the capping domain (Thr-A234 to Thr-A244 in quinol:fumarate reductase) begins at the interdomain hinge and covers the active site. Amino acids of this loop interact with both the substrate and a proton shuttle, potentially coordinating substrate binding and the proton shuttle protonation state. To assess the loop's role in catalysis, two threonine residues were mutated to alanine: QFR Thr-A244 (act-T; Thr-A254 in SQR), which hydrogen-bonds to the substrate at the active site, and QFR Thr-A234 (hinge-T; Thr-A244 in SQR), which is located at the hinge and hydrogen-bonds the proton shuttle. Both mutations impair catalysis and decrease substrate binding. The crystal structure of the hinge-T mutation reveals a reorientation between the FAD-binding and capping domains that accompanies proton shuttle alteration. Taken together, hydrogen bonding from act-T to substrate may coordinate with interdomain motions to twist the double bond of fumarate and introduce the strain important for attaining the transition state.
- Department of Pharmacology, Vanderbilt University, Nashville, TN 37232, USA.
Organizational Affiliation: