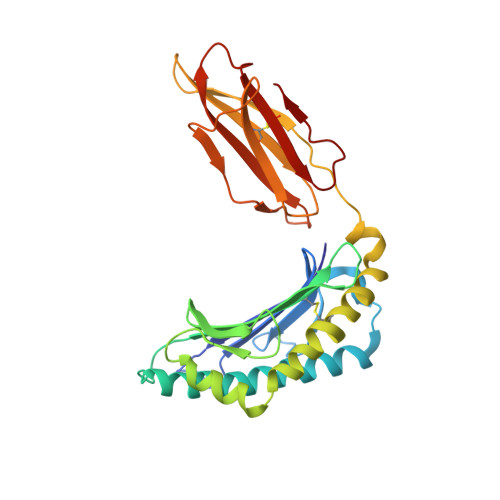
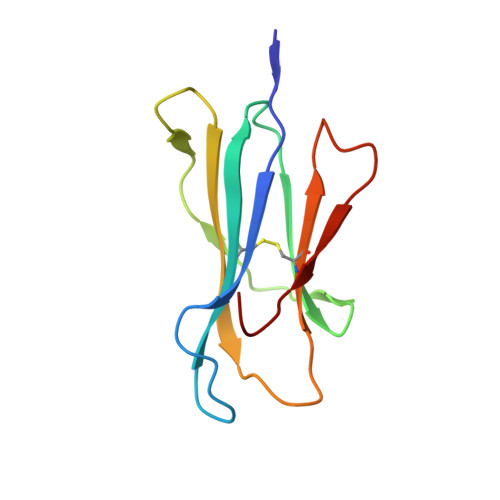
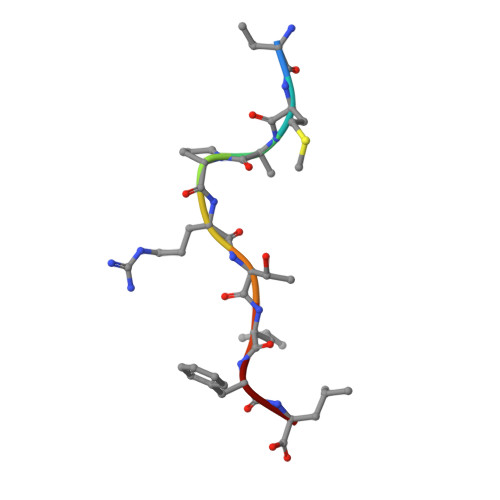
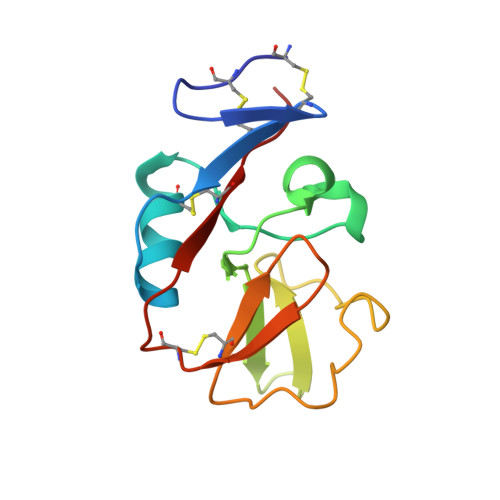
Structural basis for NKG2A/CD94 recognition of HLA-E.
Kaiser, B.K., Pizarro, J.C., Kerns, J., Strong, R.K.(2008) Proc Natl Acad Sci U S A 105: 6696-6701
- PubMed: 18448674
- DOI: https://doi.org/10.1073/pnas.0802736105
- Primary Citation of Related Structures:
3CII - PubMed Abstract:
The NKG2x/CD94 (x = A, C, E) natural killer-cell receptors perform an important role in immunosurveillance by binding to HLA-E complexes that exclusively present peptides derived from MHC class I leader sequences, thereby monitoring MHC class I expression. We have determined the crystal structure of the NKG2A/CD94/HLA-E complex at 4.4-A resolution, revealing two critical aspects of this interaction. First, the C-terminal region of the peptide, which displays the most variability among class I leader sequences, interacts entirely with CD94, the invariant component of these receptors. Second, residues 167-170 of NKG2A/C account for the approximately 6-fold-higher affinity of the inhibitory NKG2A/CD94 receptor compared to its activating NKG2C/CD94 counterpart. These residues do not contact HLA-E or peptide directly but instead form part of the heterodimer interface with CD94. An evolutionary analysis across primates reveals that whereas CD94 is evolving under purifying selection, both NKG2A and NKG2C are evolving under positive selection. Specifically, residues at the CD94 interface have evolved under positive selection, suggesting that the evolution of these genes is driven by an interaction with pathogen-derived ligands. Consistent with this possibility, we show that NKG2C/CD94, but not NKG2A/CD94, weakly but specifically binds to the CMV MHC-homologue UL18. Thus, the evolution of the NKG2x/CD94 family of receptors has likely been shaped both by the need to bind the invariant HLA-E ligand and the need to avoid subversion by pathogen-derived decoys.
- Division of Basic Sciences, Fred Hutchinson Cancer Research Center, 1100 Fairview Avenue North, Seattle, WA 98109, USA.
Organizational Affiliation: