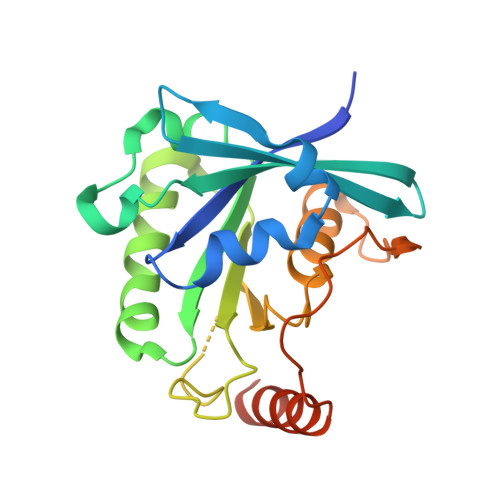
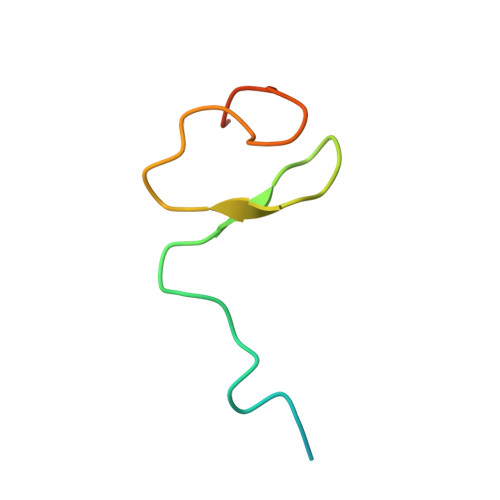
The Crystal Structure of the Ran-Nup153ZnF2 Complex: a General Ran Docking Site at the Nuclear Pore Complex
Schrader, N., Koerner, C., Koessmeier, K., Bangert, J.A., Wittinghofer, A., Stoll, R., Vetter, I.R.(2008) Structure 16: 1116-1125
- PubMed: 18611384
- DOI: https://doi.org/10.1016/j.str.2008.03.014
- Primary Citation of Related Structures:
2K0C, 3CH5 - PubMed Abstract:
Nucleoporin (Nup) 153 is a highly mobile, multifunctional, and essential nuclear pore protein. It contains four zinc finger motifs that are thought to be crucial for the regulation of transport-receptor/cargo interactions via their binding to the small guanine nucleotide binding protein, Ran. We found this interaction to be independent of the phoshorylation state of the nucleotide. Ran binds with the highest affinity to the second zinc finger motif of Nup153 (Nup153ZnF2). Here we present the crystal structure of this complex, revealing a new type of Ran-Ran interaction partner interface together with the solution structure of Nup153ZnF2. According to our complex structure, Nup153ZnF2 binding to Ran excludes the formation of a Ran-importin-beta complex. This finding suggests a local Nup153-mediated Ran reservoir at the nucleoplasmic distal ring of the nuclear pore, where nucleotide exchange may take place in a ternary Nup153-Ran-RCC1 complex, so that import complexes are efficiently terminated.
- Max-Planck-Institut für Molekulare Physiologie, Abteilung Strukturelle Biologie, Otto-Hahn-Strasse 11, 44227 Dortmund, Germany.
Organizational Affiliation: