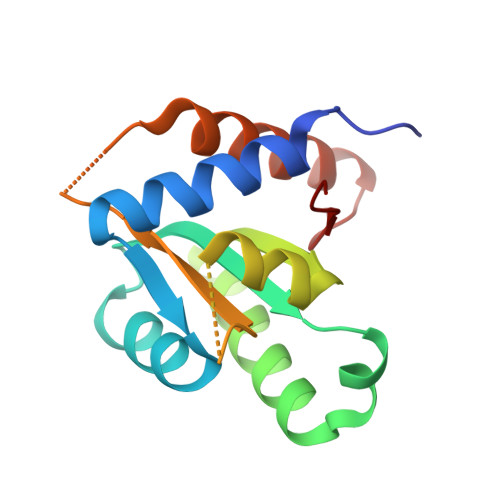
The crystal structure and dimerization interface of GADD45gamma.
Schrag, J.D., Jiralerspong, S., Banville, M., Jaramillo, M.L., O'Connor-McCourt, M.D.(2008) Proc Natl Acad Sci U S A 105: 6566-6571
- PubMed: 18445651
- DOI: https://doi.org/10.1073/pnas.0800086105
- Primary Citation of Related Structures:
3CG6 - PubMed Abstract:
Gadd45 proteins are recognized as tumor and autoimmune suppressors whose expression can be induced by genotoxic stresses. These proteins are involved in cell cycle control, growth arrest, and apoptosis through interactions with a wide variety of binding partners. We report here the crystal structure of Gadd45gamma, which reveals a fold comprising an alphabetaalpha sandwich with a central five-stranded mixed beta-sheet with alpha-helices packed on either side. Based on crystallographic symmetry we identified the dimer interface of Gadd45gamma dimers by generating point mutants that compromised dimerization while leaving the tertiary structure of the monomer intact. The dimer interface comprises a four-helix bundle involving residues that are the most highly conserved among Gadd45 isoforms. Cell-based assays using these point mutants demonstrate that dimerization is essential for growth inhibition. This structural information provides a new context for evaluation of the plethora of protein-protein interactions that govern the many functions of the Gadd45 family of proteins.
- Biotechnology Research Institute, National Research Council Canada, 6100 Royalmount Avenue, Montreal, QC, Canada. joe.schrag@nrc-cnrc.gc.ca
Organizational Affiliation: