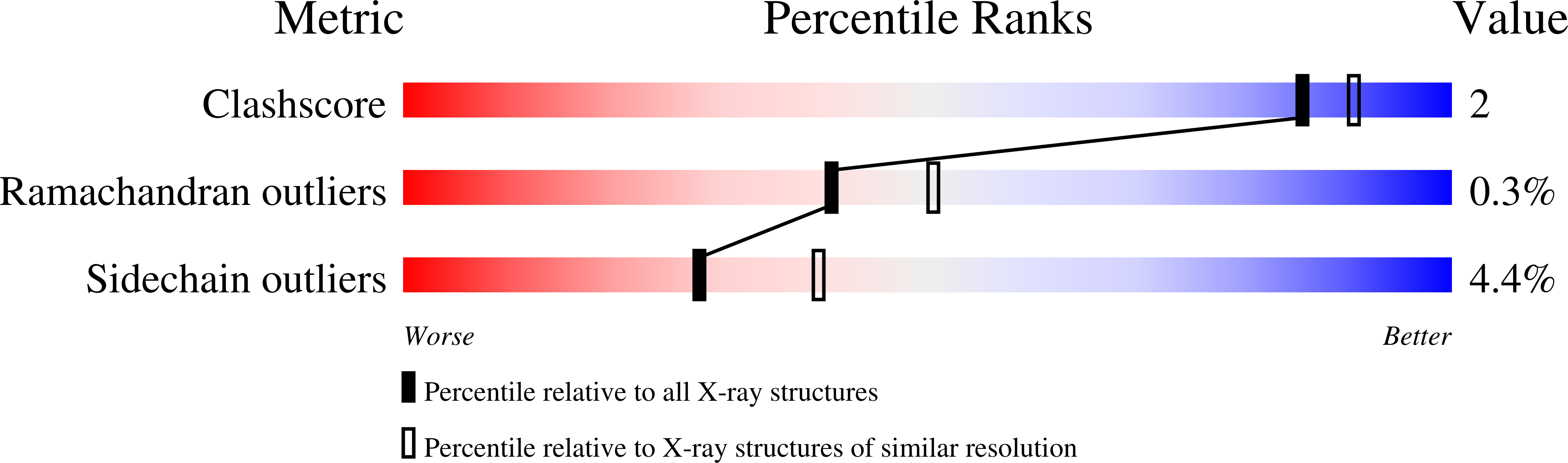Altering substrate specificity at the heme edge of cytochrome c peroxidase.
Wilcox, S.K., Jensen, G.M., Fitzgerald, M.M., McRee, D.E., Goodin, D.B.(1996) Biochemistry 35: 4858-4866
- PubMed: 8664277
- DOI: https://doi.org/10.1021/bi952929f
- Primary Citation of Related Structures:
3CCX, 4CCX - PubMed Abstract:
Two mutants of cytochrome c peroxidase (CCP) are reported which exhibit unique specificities toward oxidation of small substrates. Ala-147 in CCP is located near the delta-meso edge of the heme and along the solvent access channel through which H2O2 is thought to approach the active site. This residue was replaced with Met and Tyr to investigate the hypothesis that small molecule substrates are oxidized at the exposed delta-meso edge of the heme. X-ray crystallographic analyses confirm that the side chains of A147M and A147Y are positioned over the delta-meso heme position and might therefore modify small molecule access to the oxidized heme cofactor. Steady-state kinetic measurements show that cytochrome c oxidation is enhanced 3-fold for A147Y relative to wild type, while small molecule oxidation is altered to varying degrees depending on the substrate and mutant. For example, oxidation of phenols by A147Y is reduced to less than 20% relative to the wild-type enzyme, while Vmax/e for oxidation of other small molecules is less affected by either mutation. However, the "specificity" of aniline oxidation by A147M, i.e., (Vmax/e)/Km, is 43-fold higher than in wild-type enzyme, suggesting that a specific interaction for aniline has been introduced by the mutation. Stopped-flow kinetic data show that the restricted heme access in A147Y or A147M slows the reaction between the enzyme and H202, but not to an extent that it becomes rate limiting for the oxidation of the substrates examined. The rate constant for compound ES formation with A147Y is 2.5 times slower than wild-type CCP. These observations strongly support the suggestion that small molecule oxidations occur at sites on the enzyme distinct from those utilized by cytochrome c and that the specificity of small molecule oxidation can be significantly modulated by manipulating access to the heme edge. The results help to define the role of alternative electron transfer pathways in cytochrome c peroxidase and may have useful applications in improving the specificity of peroxidase with engineered function.
- Department of Molecular Biology, The Scripps Research Institute, La Jolla, Calfornia 92037, USA.
Organizational Affiliation: