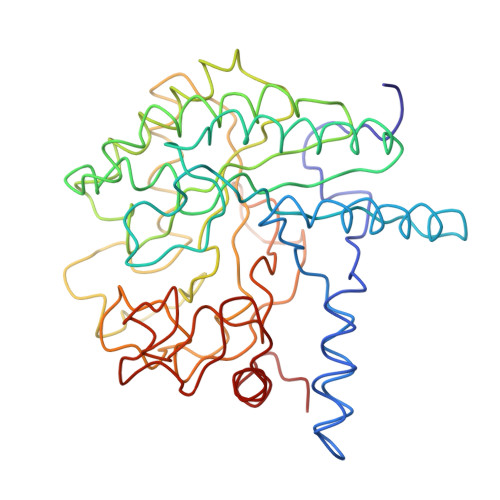
Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei.
Rouvinen, J., Bergfors, T., Teeri, T., Knowles, J.K., Jones, T.A.(1990) Science 249: 380-386
- PubMed: 2377893
- DOI: https://doi.org/10.1126/science.2377893
- Primary Citation of Related Structures:
3CBH - PubMed Abstract:
The enzymatic degradation of cellulose is an important process, both ecologically and commercially. The three-dimensional structure of a cellulase, the enzymatic core of CBHII from the fungus Trichoderma reesei reveals an alpha-beta protein with a fold similar to but different from the widely occurring barrel topology first observed in triose phosphate isomerase. The active site of CBHII is located at the carboxyl-terminal end of a parallel beta barrel, in an enclosed tunnel through which the cellulose threads. Two aspartic acid residues, located in the center of the tunnel are the probable catalytic residues.
- Department of Molecular Biology, BMC, Uppsala, Sweden.
Organizational Affiliation: